Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003821A_C01 KMC003821A_c01
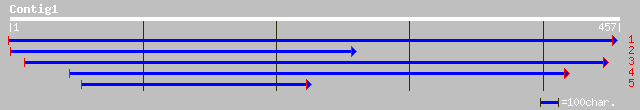
(455 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_348558.1| Hypothetical protein [Clostridium acetobutylicu... 33 1.8
ref|NP_658776.1| DUF214, Predicted permease [Bacillus anthracis ... 32 4.0
ref|NP_813280.1| conserved hypothetical protein [Bacteroides the... 32 4.0
ref|NP_113352.1| hypothetical protein [Guillardia theta] gi|2539... 31 5.2
ref|NP_147145.1| ferredoxin [Aeropyrum pernix] gi|7521058|pir||G... 30 8.8
>ref|NP_348558.1| Hypothetical protein [Clostridium acetobutylicum]
gi|25500650|pir||G97138 hypothetical protein CAC1936
[imported] - Clostridium acetobutylicum
gi|15024916|gb|AAK79898.1|AE007699_8 Hypothetical
protein [Clostridium acetobutylicum]
Length = 229
Score = 32.7 bits (73), Expect = 1.8
Identities = 19/75 (25%), Positives = 34/75 (45%), Gaps = 10/75 (13%)
Frame = +1
Query: 226 YEARNQI*EDTQYCVLRLYFENSKLWTVIGHLRTSLDTCLAPAQTDVSTHFEKKCMEF-- 399
+E +Q+ +C +R+Y E+ ++W + T +T Q S F++ C F
Sbjct: 48 WEREHQLVNGNLFCTVRVYDEDKQMWISKQDVGTESNTEKEKGQ--ASDSFKRACFNFGI 105
Query: 400 --------FLWIGMK 420
F+WIG+K
Sbjct: 106 GRELYTSPFIWIGLK 120
>ref|NP_658776.1| DUF214, Predicted permease [Bacillus anthracis A2012]
Length = 619
Score = 31.6 bits (70), Expect = 4.0
Identities = 18/54 (33%), Positives = 31/54 (57%), Gaps = 4/54 (7%)
Frame = -3
Query: 252 LLYLIPCLV----YLLGLGNFGTISYVPFFSIFFVMIIFV**EEVVFVISLLLY 103
L++L P +V L+G+ FG I P+F I+ ++IFV + + I++ LY
Sbjct: 557 LVFLFPAIVGIAHVLVGMNIFGFILIDPYFRIWVPIVIFVVIYTIYYFITVQLY 610
>ref|NP_813280.1| conserved hypothetical protein [Bacteroides thetaiotaomicron
VPI-5482] gi|29341688|gb|AAO79474.1| conserved
hypothetical protein [Bacteroides thetaiotaomicron
VPI-5482]
Length = 193
Score = 31.6 bits (70), Expect = 4.0
Identities = 16/55 (29%), Positives = 25/55 (45%)
Frame = +1
Query: 157 HDKKY*KKWNITYGTKISQTQQIYEARNQI*EDTQYCVLRLYFENSKLWTVIGHL 321
+D+K W G + Q I+ I DT Y Y++ KLW +IG++
Sbjct: 55 YDRKKPSYWAFEKGVYLEQFDSIFNIEASIKADTAY-----YYDKQKLWKLIGNV 104
>ref|NP_113352.1| hypothetical protein [Guillardia theta] gi|25396799|pir||E90097
hypothetical protein orf1019 [imported] - Guillardia
theta nucleomorph gi|13794533|gb|AAK39908.1|AF165818_116
hypothetical protein [Guillardia theta]
Length = 1019
Score = 31.2 bits (69), Expect = 5.2
Identities = 18/73 (24%), Positives = 34/73 (45%), Gaps = 10/73 (13%)
Frame = -2
Query: 193 KLCSIFFNIFCHD----------HFCVMRRSSLCNFITVVF*TADNELMIAVAFPRCKTH 44
K +I NIFC + HFC+++ S +F++ +F N + + + K H
Sbjct: 218 KFFNIITNIFCQNFCLKFDIFFLHFCLLKNSVYLSFMSRIFYKLQNSVNYFSSKLKMKKH 277
Query: 43 IKRDNFESCYIFF 5
+ ++ F+ I F
Sbjct: 278 LYKNYFKYSNILF 290
>ref|NP_147145.1| ferredoxin [Aeropyrum pernix] gi|7521058|pir||G72722 probable
ferredoxin APE0320 - Aeropyrum pernix (strain K1)
gi|5103959|dbj|BAA79275.1| 115aa long hypothetical
ferredoxin [Aeropyrum pernix]
Length = 115
Score = 30.4 bits (67), Expect = 8.8
Identities = 17/69 (24%), Positives = 29/69 (41%)
Frame = +1
Query: 13 YSNFQSCLFLCVFCSEEMPQQSLTHYQQFKIQQ**NYKDYFFSSHKNDHDKKY*KKWNIT 192
+S +SC C++ EE P++ T + Y N+ +KK ++W
Sbjct: 56 FSCIESCPVNCIYKVEEAPEELKTEKAGW----------YRLGRELNEEEKKAFEEWRQK 105
Query: 193 YGTKISQTQ 219
YG K+ Q
Sbjct: 106 YGVKVDPVQ 114
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 402,571,259
Number of Sequences: 1393205
Number of extensions: 8619738
Number of successful extensions: 20917
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 20364
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20908
length of database: 448,689,247
effective HSP length: 112
effective length of database: 292,650,287
effective search space used: 11413361193
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)