Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
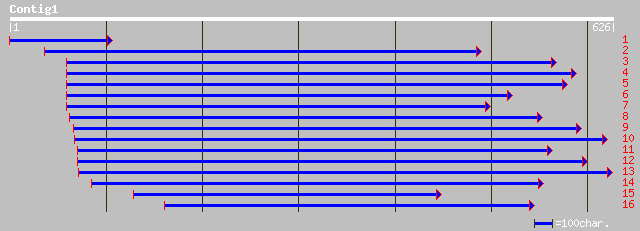
Query= KMC003816A_C01 KMC003816A_c01
(623 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||S41880 1-aminocyclopropane-1-carboxylate oxidase [similarit... 225 4e-58
prf||1909340A Pch313 protein 225 4e-58
sp|Q9MB94|ACCO_PRUMU 1-aminocyclopropane-1-carboxylate oxidase (... 220 1e-56
dbj|BAA94601.1| 1-aminocyclopropane-1-carboxylate oxidase [Popul... 218 5e-56
dbj|BAB83762.1| 1-aminocyclopropane-1-carboxylic acid oxidase [P... 215 4e-55
>pir||S41880 1-aminocyclopropane-1-carboxylate oxidase [similarity] - peach
gi|452671|emb|CAA54449.1|
1-aminocyclopropane-1-carboxylate oxidase [Prunus
persica] gi|3510500|gb|AAC33524.1|
1-aminocyclopropane-1-carboxylate oxidase; ACC oxidase
[Prunus armeniaca] gi|7108577|gb|AAF36483.1|AF129073_1
1-aminocyclopropane-1-carboxylate oxidase [Prunus
persica] gi|16588828|gb|AAL26910.1|AF319166_1
1-aminocyclopropane 1-carboxylic acid oxidase [Prunus
persica]
Length = 319
Score = 225 bits (573), Expect = 4e-58
Identities = 109/123 (88%), Positives = 117/123 (94%), Gaps = 1/123 (0%)
Frame = -1
Query: 623 LLKDGEWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD 444
LLKDG+W+DVPPMRHSIV+NLGDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD
Sbjct: 197 LLKDGQWIDVPPMRHSIVINLGDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD 256
Query: 443 AVIYPAPELLEKEAEEKNQLYPKFVFEDYMKLYAGLKFDAKEPRFEALKA-SPNVTLGPI 267
AVIYPAP L+EKEAEEKNQ+YPKFVFEDYMKLYAGLKF KEPRFEA+KA N++LGPI
Sbjct: 257 AVIYPAPTLVEKEAEEKNQVYPKFVFEDYMKLYAGLKFQPKEPRFEAMKAVETNISLGPI 316
Query: 266 ATA 258
ATA
Sbjct: 317 ATA 319
>prf||1909340A Pch313 protein
Length = 319
Score = 225 bits (573), Expect = 4e-58
Identities = 109/123 (88%), Positives = 117/123 (94%), Gaps = 1/123 (0%)
Frame = -1
Query: 623 LLKDGEWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD 444
LLKDG+W+DVPPMRHSIV+NLGDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD
Sbjct: 197 LLKDGQWIDVPPMRHSIVINLGDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD 256
Query: 443 AVIYPAPELLEKEAEEKNQLYPKFVFEDYMKLYAGLKFDAKEPRFEALKA-SPNVTLGPI 267
AVIYPAP L+EKEAEEKNQ+YPKFVFEDYMKLYAGLKF KEPRFEA+KA N++LGPI
Sbjct: 257 AVIYPAPTLVEKEAEEKNQVYPKFVFEDYMKLYAGLKFQPKEPRFEAMKAVETNISLGPI 316
Query: 266 ATA 258
ATA
Sbjct: 317 ATA 319
>sp|Q9MB94|ACCO_PRUMU 1-aminocyclopropane-1-carboxylate oxidase (ACC oxidase)
(Ethylene-forming enzyme) (EFE)
gi|6906698|dbj|BAA90550.1| ACC oxidase [Prunus mume]
Length = 319
Score = 220 bits (561), Expect = 1e-56
Identities = 107/123 (86%), Positives = 116/123 (93%), Gaps = 1/123 (0%)
Frame = -1
Query: 623 LLKDGEWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD 444
LLKDG+W+DVPPMRHSIV+NLGDQLEVITNGKY+SVEHRVIAQTDGTRMSIASFYNPGSD
Sbjct: 197 LLKDGQWIDVPPMRHSIVINLGDQLEVITNGKYRSVEHRVIAQTDGTRMSIASFYNPGSD 256
Query: 443 AVIYPAPELLEKEAEEKNQLYPKFVFEDYMKLYAGLKFDAKEPRFEALKA-SPNVTLGPI 267
AVIYPAP L+EKEAEEKNQ+YPKFVFEDYMKLYAGLKF KEPRFEA+KA N++L PI
Sbjct: 257 AVIYPAPTLVEKEAEEKNQVYPKFVFEDYMKLYAGLKFQPKEPRFEAMKAVETNISLVPI 316
Query: 266 ATA 258
ATA
Sbjct: 317 ATA 319
>dbj|BAA94601.1| 1-aminocyclopropane-1-carboxylate oxidase [Populus euramericana]
Length = 319
Score = 218 bits (555), Expect = 5e-56
Identities = 106/123 (86%), Positives = 115/123 (93%), Gaps = 1/123 (0%)
Frame = -1
Query: 623 LLKDGEWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD 444
LLKDG+W+DVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTDGTRMS+ASFYNPGS+
Sbjct: 197 LLKDGQWIDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTDGTRMSVASFYNPGSE 256
Query: 443 AVIYPAPELLEKEAEEKNQLYPKFVFEDYMKLYAGLKFDAKEPRFEALKA-SPNVTLGPI 267
AVIYPAP L+EKEAEEK ++YPKFVF+DYMKLYAGLKF AKEPRFEA+KA V LGPI
Sbjct: 257 AVIYPAPALVEKEAEEKKKVYPKFVFDDYMKLYAGLKFQAKEPRFEAMKAVETTVNLGPI 316
Query: 266 ATA 258
ATA
Sbjct: 317 ATA 319
>dbj|BAB83762.1| 1-aminocyclopropane-1-carboxylic acid oxidase [Phaseolus lunatus]
Length = 315
Score = 215 bits (547), Expect = 4e-55
Identities = 109/122 (89%), Positives = 111/122 (90%)
Frame = -1
Query: 623 LLKDGEWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD 444
LLKD +WVDVPPMRHSIVVN+GDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD
Sbjct: 197 LLKDDQWVDVPPMRHSIVVNIGDQLEVITNGKYKSVEHRVIAQTDGTRMSIASFYNPGSD 256
Query: 443 AVIYPAPELLEKEAEEKNQLYPKFVFEDYMKLYAGLKFDAKEPRFEALKASPNVTLGPIA 264
AVIYPAPELLEKEAEEK QLYPKFVFEDYMKLYA LKF AKEPRFEA KAS PIA
Sbjct: 257 AVIYPAPELLEKEAEEKAQLYPKFVFEDYMKLYAKLKFQAKEPRFEAFKAS---NFDPIA 313
Query: 263 TA 258
TA
Sbjct: 314 TA 315
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 547,791,045
Number of Sequences: 1393205
Number of extensions: 11425799
Number of successful extensions: 28891
Number of sequences better than 10.0: 629
Number of HSP's better than 10.0 without gapping: 28066
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28765
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25301904073
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)