Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003815A_C01 KMC003815A_c01
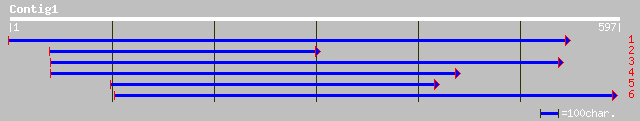
(596 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN87739.1| Unknown protein [Oryza sativa (japonica cultivar-... 84 1e-15
pir||T03812 hypothetical protein (clone NF22) - common tobacco g... 79 3e-14
emb|CAD39524.1| OSJNBa0027O01.12 [Oryza sativa (japonica cultiva... 77 1e-13
ref|NP_193178.1| elicitor like protein; protein id: At4g14420.1,... 66 3e-10
gb|AAC77929.1| similar to Nicotiana HR lesion-inducing ORF [Medi... 65 5e-10
>gb|AAN87739.1| Unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 157
Score = 84.3 bits (207), Expect = 1e-15
Identities = 40/53 (75%), Positives = 47/53 (88%), Gaps = 1/53 (1%)
Frame = -3
Query: 594 DFYNFDHEDKEFAQLFIEFTQNMALFGALLFFIGMKNSIPRRQ-PKKAPKTKT 439
DFYN+D E EF QLF++FTQN+ALFGALLFF+GMKNSIP+RQ KKAPK+KT
Sbjct: 104 DFYNYDMEKSEFVQLFMKFTQNLALFGALLFFLGMKNSIPKRQAKKKAPKSKT 156
>pir||T03812 hypothetical protein (clone NF22) - common tobacco
gi|1762945|gb|AAC49975.1| ORF; able to induce HR-like
lesions [Nicotiana tabacum]
Length = 157
Score = 79.3 bits (194), Expect = 3e-14
Identities = 38/53 (71%), Positives = 45/53 (84%), Gaps = 1/53 (1%)
Frame = -3
Query: 594 DFYNFDHEDKEFAQLFIEFTQNMALFGALLFFIGMKNSIPRR-QPKKAPKTKT 439
DFYN+D + KEF QLF +F+QN+AL GALLFFIGMKNS+PRR KKAPK+KT
Sbjct: 104 DFYNYDVDKKEFVQLFFKFSQNLALLGALLFFIGMKNSMPRRSSKKKAPKSKT 156
>emb|CAD39524.1| OSJNBa0027O01.12 [Oryza sativa (japonica cultivar-group)]
Length = 157
Score = 77.4 bits (189), Expect = 1e-13
Identities = 36/53 (67%), Positives = 45/53 (83%), Gaps = 1/53 (1%)
Frame = -3
Query: 594 DFYNFDHEDKEFAQLFIEFTQNMALFGALLFFIGMKNSIPRRQPK-KAPKTKT 439
DFYN++ E +F QLF++F+QN+ALFGALLFF+GMKNSIPRR K +A KTKT
Sbjct: 104 DFYNYEMESSQFVQLFVKFSQNLALFGALLFFLGMKNSIPRRHSKRRAGKTKT 156
>ref|NP_193178.1| elicitor like protein; protein id: At4g14420.1, supported by cDNA:
1718., supported by cDNA: gi_15010599, supported by
cDNA: gi_19699241 [Arabidopsis thaliana]
gi|7485063|pir||C71406 hypothetical protein -
Arabidopsis thaliana gi|2244798|emb|CAB10221.1| elicitor
like protein [Arabidopsis thaliana]
gi|7268148|emb|CAB78484.1| elicitor like protein
[Arabidopsis thaliana] gi|15010600|gb|AAK73959.1|
AT4g14420/dl3250c [Arabidopsis thaliana]
gi|19699242|gb|AAL90987.1| AT4g14420/dl3250c
[Arabidopsis thaliana] gi|21553825|gb|AAM62918.1|
elicitor like protein [Arabidopsis thaliana]
Length = 158
Score = 66.2 bits (160), Expect = 3e-10
Identities = 34/53 (64%), Positives = 40/53 (75%), Gaps = 2/53 (3%)
Frame = -3
Query: 594 DFYNFDHEDKEFAQLFIEFTQNMALFGALLFFIGMKNSIP--RRQPKKAPKTK 442
DFYN+D + KEF QLF +FTQ++AL G LLFFIGMKNS R+ KKAPK K
Sbjct: 104 DFYNYDVDRKEFGQLFSKFTQSLALLGGLLFFIGMKNSRKHGRQLRKKAPKAK 156
>gb|AAC77929.1| similar to Nicotiana HR lesion-inducing ORF [Medicago sativa]
Length = 130
Score = 65.5 bits (158), Expect = 5e-10
Identities = 34/52 (65%), Positives = 37/52 (70%), Gaps = 1/52 (1%)
Frame = -3
Query: 594 DFYNFDHEDKEFAQLFIEFTQNMALFGALLFFIGMKNSIPRRQ-PKKAPKTK 442
DFYN+ E+ L EF QN ALFGALLFFIGMKNSIPR+Q KK PK K
Sbjct: 77 DFYNYRPTKPEYGLLLNEFIQNAALFGALLFFIGMKNSIPRKQLRKKTPKAK 128
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 523,992,788
Number of Sequences: 1393205
Number of extensions: 11532085
Number of successful extensions: 35201
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 33454
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35152
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23140425222
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)