Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003801A_C01 KMC003801A_c01
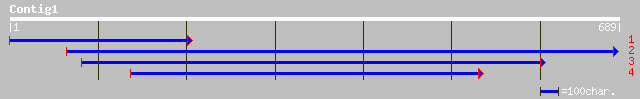
(685 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG29593.1|AF196286_1 Ser/Thr specific protein phosphatase 2A... 208 6e-53
ref|NP_189208.1| protein phosphatase 2A 65 kDa regulatory subuni... 197 9e-50
pir||S51808 phosphoprotein phosphatase 2A 65K regulatory chain h... 197 9e-50
pir||S51809 phosphoprotein phosphatase 2A 65K regulatory chain h... 193 2e-48
ref|NP_172790.2| protein phosphatase 2A regulatory subunit, puta... 193 2e-48
>gb|AAG29593.1|AF196286_1 Ser/Thr specific protein phosphatase 2A A regulatory subunit alpha
isoform [Medicago sativa subsp. x varia]
Length = 585
Score = 208 bits (529), Expect = 6e-53
Identities = 104/114 (91%), Positives = 110/114 (96%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+IIPQVL+MI NPHYLYRMTILRAISLLAPVMGSE+TCSKLLP VV+ASKDRVPNIKFN
Sbjct: 472 QHIIPQVLEMITNPHYLYRMTILRAISLLAPVMGSEVTCSKLLPAVVAASKDRVPNIKFN 531
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMST 343
VAKVLESIFPI+DQSVVEKTIRPCLVEL EDPDVDVRFFSN ALQAIDHVMMS+
Sbjct: 532 VAKVLESIFPIVDQSVVEKTIRPCLVELGEDPDVDVRFFSNQALQAIDHVMMSS 585
Score = 57.0 bits (136), Expect = 2e-07
Identities = 28/97 (28%), Positives = 55/97 (55%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+P V ++ ++ R + I +APV+G E T +LL + +S KD P+++ N
Sbjct: 316 QHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKEATIEQLLHIFLSLLKDEFPDVRIN 375
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 394
+ L+ + ++ ++ +++ P +VEL+ED VR
Sbjct: 376 IISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 412
Score = 38.9 bits (89), Expect = 0.067
Identities = 23/96 (23%), Positives = 43/96 (43%)
Frame = -2
Query: 681 NIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFNV 502
+I+P +++ + + R + + L +G E T ++L+P V +D ++
Sbjct: 239 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRAELVPAYVRLLRDNEAEVRIAA 298
Query: 501 AKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 394
A + IL + + I PC+ ELS D VR
Sbjct: 299 AGKVTKFCRILSPDLAIQHILPCVKELSSDSSQHVR 334
Score = 34.7 bits (78), Expect = 1.3
Identities = 26/113 (23%), Positives = 53/113 (46%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+++P ++++ + H+ R+ I+ I LLA +G KL + + +D+V +I+
Sbjct: 394 QSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLQDKVHSIREA 453
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMS 346
A L+ + + I P ++E+ +P R A+ + VM S
Sbjct: 454 AANNLKRLAEEFGPEWAMQHIIPQVLEMITNPHYLYRMTILRAISLLAPVMGS 506
Score = 32.0 bits (71), Expect = 8.1
Identities = 18/109 (16%), Positives = 47/109 (42%)
Frame = -2
Query: 678 IIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFNVA 499
++P + ++ + R+ ++ ++ ++ +LP V S D +++ +A
Sbjct: 279 LVPAYVRLLRDNEAEVRIAAAGKVTKFCRILSPDLAIQHILPCVKELSSDSSQHVRSALA 338
Query: 498 KVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVM 352
V+ + P+L + + + + L +D DVR L ++ V+
Sbjct: 339 SVIMGMAPVLGKEATIEQLLHIFLSLLKDEFPDVRINIISKLDQVNQVI 387
>ref|NP_189208.1| protein phosphatase 2A 65 kDa regulatory subunit; protein id:
At3g25800.1 [Arabidopsis thaliana]
gi|7939566|dbj|BAA95767.1| protein phosphotase 2a 65kd
regulatory subunit [Arabidopsis thaliana]
gi|27311765|gb|AAO00848.1| protein phosphatase 2A 65 kDa
regulatory subunit [Arabidopsis thaliana]
Length = 587
Score = 197 bits (502), Expect = 9e-50
Identities = 97/114 (85%), Positives = 111/114 (97%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+PQVL+M+NNPHYLYRMTILRA+SLLAPVMGSEITCSKLLPVV++ASKDRVPNIKFN
Sbjct: 474 QHIVPQVLEMVNNPHYLYRMTILRAVSLLAPVMGSEITCSKLLPVVMTASKDRVPNIKFN 533
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMST 343
VAKVL+S+ PI+DQSVVEKTIRP LVELSEDPDVDVRFF+N ALQ+ID+VMMS+
Sbjct: 534 VAKVLQSLIPIVDQSVVEKTIRPGLVELSEDPDVDVRFFANQALQSIDNVMMSS 587
Score = 58.5 bits (140), Expect = 8e-08
Identities = 28/97 (28%), Positives = 55/97 (55%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+P V ++ ++ R + I +APV+G + T LLP+ +S KD P+++ N
Sbjct: 318 QHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDVRLN 377
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 394
+ L+ + ++ ++ +++ P +VEL+ED VR
Sbjct: 378 IISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 414
Score = 42.0 bits (97), Expect = 0.008
Identities = 24/97 (24%), Positives = 45/97 (45%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+P +++ + + R + + L +G E T ++L+P V +D ++
Sbjct: 240 QHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIA 299
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 394
A + IL+ + + I PC+ ELS D VR
Sbjct: 300 AAGKVTKFCRILNPEIAIQHILPCVKELSSDSSQHVR 336
Score = 37.0 bits (84), Expect = 0.25
Identities = 21/109 (19%), Positives = 48/109 (43%)
Frame = -2
Query: 678 IIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFNVA 499
++P + ++ + R+ ++ ++ EI +LP V S D +++ +A
Sbjct: 281 LVPAYVRLLRDNEAEVRIAAAGKVTKFCRILNPEIAIQHILPCVKELSSDSSQHVRSALA 340
Query: 498 KVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVM 352
V+ + P+L + + + P + L +D DVR L ++ V+
Sbjct: 341 SVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDVRLNIISKLDQVNQVI 389
Score = 34.7 bits (78), Expect = 1.3
Identities = 26/113 (23%), Positives = 53/113 (46%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+++P ++++ + H+ R+ I+ I LLA +G KL + + +D+V +I+
Sbjct: 396 QSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLQDKVHSIRDA 455
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMS 346
A L+ + + I P ++E+ +P R A+ + VM S
Sbjct: 456 AANNLKRLAEEFGPEWAMQHIVPQVLEMVNNPHYLYRMTILRAVSLLAPVMGS 508
>pir||S51808 phosphoprotein phosphatase 2A 65K regulatory chain homolog pDF1 -
Arabidopsis thaliana gi|683502|emb|CAA57528.1| protein
phosphatase 2A 65 kDa regulatory subunit [Arabidopsis
thaliana]
Length = 587
Score = 197 bits (502), Expect = 9e-50
Identities = 97/114 (85%), Positives = 111/114 (97%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+PQVL+M+NNPHYLYRMTILRA+SLLAPVMGSEITCSKLLPVV++ASKDRVPNIKFN
Sbjct: 474 QHIVPQVLEMVNNPHYLYRMTILRAVSLLAPVMGSEITCSKLLPVVMTASKDRVPNIKFN 533
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMST 343
VAKVL+S+ PI+DQSVVEKTIRP LVELSEDPDVDVRFF+N ALQ+ID+VMMS+
Sbjct: 534 VAKVLQSLIPIVDQSVVEKTIRPGLVELSEDPDVDVRFFANQALQSIDNVMMSS 587
Score = 58.5 bits (140), Expect = 8e-08
Identities = 28/97 (28%), Positives = 55/97 (55%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+P V ++ ++ R + I +APV+G + T LLP+ +S KD P+++ N
Sbjct: 318 QDILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDVRLN 377
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 394
+ L+ + ++ ++ +++ P +VEL+ED VR
Sbjct: 378 IISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 414
Score = 42.4 bits (98), Expect = 0.006
Identities = 24/97 (24%), Positives = 45/97 (45%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+P +++ + + R + + L +G E T ++L+P V +D ++
Sbjct: 240 QHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIA 299
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 394
A + IL+ + + I PC+ ELS D VR
Sbjct: 300 AAGKVTKFCRILNPEIAIQDILPCVKELSSDSSQHVR 336
Score = 37.0 bits (84), Expect = 0.25
Identities = 21/109 (19%), Positives = 48/109 (43%)
Frame = -2
Query: 678 IIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFNVA 499
++P + ++ + R+ ++ ++ EI +LP V S D +++ +A
Sbjct: 281 LVPAYVRLLRDNEAEVRIAAAGKVTKFCRILNPEIAIQDILPCVKELSSDSSQHVRSALA 340
Query: 498 KVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVM 352
V+ + P+L + + + P + L +D DVR L ++ V+
Sbjct: 341 SVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDVRLNIISKLDQVNQVI 389
Score = 34.7 bits (78), Expect = 1.3
Identities = 26/113 (23%), Positives = 53/113 (46%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+++P ++++ + H+ R+ I+ I LLA +G KL + + +D+V +I+
Sbjct: 396 QSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLQDKVHSIRDA 455
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMS 346
A L+ + + I P ++E+ +P R A+ + VM S
Sbjct: 456 AANNLKRLAEEFGPEWAMQHIVPQVLEMVNNPHYLYRMTILRAVSLLAPVMGS 508
>pir||S51809 phosphoprotein phosphatase 2A 65K regulatory chain homolog pDF2 -
Arabidopsis thaliana (fragment)
gi|683504|emb|CAA57529.1| protein phosphatase 2A 65 kDa
regulatory subunit [Arabidopsis thaliana]
Length = 508
Score = 193 bits (491), Expect = 2e-48
Identities = 92/114 (80%), Positives = 109/114 (94%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+PQVL+MINNPHYLYRMTILRA+SLLAPVMGSEITCSKLLP V++ASKDRVPNIKFN
Sbjct: 395 QHIVPQVLEMINNPHYLYRMTILRAVSLLAPVMGSEITCSKLLPAVITASKDRVPNIKFN 454
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMST 343
VAK+++S+ PI+DQ+VVE IRPCLVELSEDPDVDVR+F+N ALQ+ID+VMMS+
Sbjct: 455 VAKMMQSLIPIVDQAVVENMIRPCLVELSEDPDVDVRYFANQALQSIDNVMMSS 508
Score = 58.5 bits (140), Expect = 8e-08
Identities = 28/97 (28%), Positives = 55/97 (55%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+P V ++ ++ R + I +APV+G + T LLP+ +S KD P+++ N
Sbjct: 239 QHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDVRLN 298
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 394
+ L+ + ++ ++ +++ P +VEL+ED VR
Sbjct: 299 IISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 335
Score = 37.0 bits (84), Expect = 0.25
Identities = 22/96 (22%), Positives = 42/96 (42%)
Frame = -2
Query: 681 NIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFNV 502
+I+P +++ + + R + + L +G E T + L+P +D ++
Sbjct: 162 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTDLVPAYARLLRDNEAEVRIAA 221
Query: 501 AKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 394
A + IL+ + + I PC+ ELS D VR
Sbjct: 222 AGKVTKFCRILNPELAIQHILPCVKELSSDSSQHVR 257
Score = 35.8 bits (81), Expect = 0.56
Identities = 20/110 (18%), Positives = 48/110 (43%)
Frame = -2
Query: 681 NIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFNV 502
+++P ++ + R+ ++ ++ E+ +LP V S D +++ +
Sbjct: 201 DLVPAYARLLRDNEAEVRIAAAGKVTKFCRILNPELAIQHILPCVKELSSDSSQHVRSAL 260
Query: 501 AKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVM 352
A V+ + P+L + + + P + L +D DVR L ++ V+
Sbjct: 261 ASVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDVRLNIISKLDQVNQVI 310
Score = 34.7 bits (78), Expect = 1.3
Identities = 26/113 (23%), Positives = 53/113 (46%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+++P ++++ + H+ R+ I+ I LLA +G KL + + +D+V +I+
Sbjct: 317 QSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDEKLGALCMQWLQDKVHSIREA 376
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMS 346
A L+ + + I P ++E+ +P R A+ + VM S
Sbjct: 377 AANNLKRLAEEFGPEWAMQHIVPQVLEMINNPHYLYRMTILRAVSLLAPVMGS 429
>ref|NP_172790.2| protein phosphatase 2A regulatory subunit, putative; protein id:
At1g13320.1, supported by cDNA: gi_20466587 [Arabidopsis
thaliana] gi|20466588|gb|AAM20611.1| protein phosphatase
2A regulatory subunit, putative [Arabidopsis thaliana]
gi|27311991|gb|AAO00961.1| protein phosphatase 2A
regulatory subunit, putative [Arabidopsis thaliana]
Length = 587
Score = 193 bits (491), Expect = 2e-48
Identities = 92/114 (80%), Positives = 109/114 (94%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+PQVL+MINNPHYLYRMTILRA+SLLAPVMGSEITCSKLLP V++ASKDRVPNIKFN
Sbjct: 474 QHIVPQVLEMINNPHYLYRMTILRAVSLLAPVMGSEITCSKLLPAVITASKDRVPNIKFN 533
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMST 343
VAK+++S+ PI+DQ+VVE IRPCLVELSEDPDVDVR+F+N ALQ+ID+VMMS+
Sbjct: 534 VAKMMQSLIPIVDQAVVENMIRPCLVELSEDPDVDVRYFANQALQSIDNVMMSS 587
Score = 58.5 bits (140), Expect = 8e-08
Identities = 28/97 (28%), Positives = 55/97 (55%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+I+P V ++ ++ R + I +APV+G + T LLP+ +S KD P+++ N
Sbjct: 318 QHILPCVKELSSDSSQHVRSALASVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDVRLN 377
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 394
+ L+ + ++ ++ +++ P +VEL+ED VR
Sbjct: 378 IISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 414
Score = 35.0 bits (79), Expect = 0.96
Identities = 22/96 (22%), Positives = 41/96 (41%)
Frame = -2
Query: 681 NIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFNV 502
+I+P +++ + + R + + L +G E T + L+P D ++
Sbjct: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTDLVPAYARLLCDNEAEVRIAA 300
Query: 501 AKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 394
A + IL+ + + I PC+ ELS D VR
Sbjct: 301 AGKVTKFCRILNPELAIQHILPCVKELSSDSSQHVR 336
Score = 34.7 bits (78), Expect = 1.3
Identities = 20/110 (18%), Positives = 48/110 (43%)
Frame = -2
Query: 681 NIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFNV 502
+++P ++ + R+ ++ ++ E+ +LP V S D +++ +
Sbjct: 280 DLVPAYARLLCDNEAEVRIAAAGKVTKFCRILNPELAIQHILPCVKELSSDSSQHVRSAL 339
Query: 501 AKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVM 352
A V+ + P+L + + + P + L +D DVR L ++ V+
Sbjct: 340 ASVIMGMAPVLGKDATIEHLLPIFLSLLKDEFPDVRLNIISKLDQVNQVI 389
Score = 34.7 bits (78), Expect = 1.3
Identities = 26/113 (23%), Positives = 53/113 (46%)
Frame = -2
Query: 684 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 505
Q+++P ++++ + H+ R+ I+ I LLA +G KL + + +D+V +I+
Sbjct: 396 QSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDEKLGALCMQWLQDKVHSIREA 455
Query: 504 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMS 346
A L+ + + I P ++E+ +P R A+ + VM S
Sbjct: 456 AANNLKRLAEEFGPEWAMQHIVPQVLEMINNPHYLYRMTILRAVSLLAPVMGS 508
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 570,695,205
Number of Sequences: 1393205
Number of extensions: 11537851
Number of successful extensions: 22789
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 21964
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22730
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30552968016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)