Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003787A_C01 KMC003787A_c01
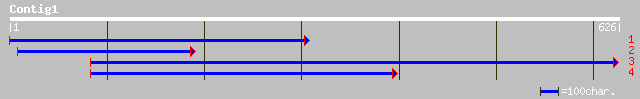
(611 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_700554.1| endonuclease, putative [Plasmodium falciparum 3... 36 0.45
pir||S36956 cytochrome-c oxidase (EC 1.9.3.1) chain III - Herpet... 35 1.0
pir||T30050 hypothetical protein K06C4.9 - Caenorhabditis elegans 34 1.3
gb|AAK56876.1|AF314019_2 NADH dehydrogenase subunit II [Rana boy... 34 1.3
ref|NP_505299.1| Putative plasma membrane membrane protein famil... 34 1.3
>ref|NP_700554.1| endonuclease, putative [Plasmodium falciparum 3D7]
gi|23494944|gb|AAN35278.1|AE014830_22 endonuclease,
putative [Plasmodium falciparum 3D7]
Length = 388
Score = 35.8 bits (81), Expect = 0.45
Identities = 26/108 (24%), Positives = 50/108 (46%), Gaps = 1/108 (0%)
Frame = -2
Query: 466 NVLSIFSLHNCKPTIKKSKVPINGGDEAVSIVQEIDDDYQTGELFSD-ACDEDNNDSTTS 290
N+ I ++ + K I KS IN D I++ ++ + ++ CD+ NN S
Sbjct: 134 NLFDILNVDSSKNNILKSNDNINVND-----TNTINNTFKKNNIVNNFLCDQKNNISNIC 188
Query: 289 RFERLKNEDMTQSLIDERGSWICFC*YGMKSLTSRTKNRAFDMLRMKN 146
+ LK ++ + ++DE G F + S+T N ++ L ++N
Sbjct: 189 EKKNLKKKNQMEIIVDEDGIHDLFKKNIFIKINSKTANDIYNYLLLEN 236
>pir||S36956 cytochrome-c oxidase (EC 1.9.3.1) chain III - Herpetomonas
mariadeanei mitochondrion gi|3559762|gb|AAD09163.1|
cytochrome oxidase subunit III [Herpetomonas
mariadeanei]
Length = 288
Score = 34.7 bits (78), Expect = 1.0
Identities = 12/43 (27%), Positives = 25/43 (57%)
Frame = +2
Query: 305 IIVFITCITKKFSSLIVIINFLHYRHCFVTTINRYLALLDCWL 433
+I+F+TC F S++ I+ LH+ H F+ ++ + C++
Sbjct: 202 VILFVTCSCGLFGSILFCIDILHFTHVFLGVFLMFICICRCFV 244
>pir||T30050 hypothetical protein K06C4.9 - Caenorhabditis elegans
Length = 819
Score = 34.3 bits (77), Expect = 1.3
Identities = 19/49 (38%), Positives = 28/49 (56%)
Frame = +3
Query: 117 VFSPFEIFFLFFILNISNALFLVLDVKLFIPY*QKQIQEPLSSMRLCVI 263
VFS F+IFFL FI+N+S L L I + I++P +S+ L +
Sbjct: 235 VFSVFKIFFLLFIINVSIILLLKYSHNERIRMTSQSIRDPRTSIMLLAV 283
>gb|AAK56876.1|AF314019_2 NADH dehydrogenase subunit II [Rana boylii]
Length = 344
Score = 34.3 bits (77), Expect = 1.3
Identities = 26/111 (23%), Positives = 57/111 (50%)
Frame = +3
Query: 258 VISSFLSLSNLDVVESLLSSSHASLKSSPV**SSSISCTIDTASSPPLIGTLLFLIVGLQ 437
++++ + +S + V + +S AS +P ++++ + A PPL G L++ L+
Sbjct: 209 IMTAAMFMSLISVSSTKMSQISASWSKTPALSTTTMLVMLSLAGLPPLTGFAPKLLITLE 268
Query: 438 LCKENIDSTLFTNMYVVVDLYSNIQTCNAPMRAYYLIAESVTWGTACSLLT 590
L K+N +TL + +++ L + +R Y+I +++ T SL+T
Sbjct: 269 LVKQN--ATLLAAIIMLISLLALF----FYLRLTYIITMTLSPNTPSSLVT 313
>ref|NP_505299.1| Putative plasma membrane membrane protein family member, with at
least 5 transmembrane domains, of bilaterial origin
[Caenorhabditis elegans]
gi|15789300|gb|AAL08038.1|U64843_8 Hypothetical protein
K06C4.17 [Caenorhabditis elegans]
Length = 365
Score = 34.3 bits (77), Expect = 1.3
Identities = 19/49 (38%), Positives = 28/49 (56%)
Frame = +3
Query: 117 VFSPFEIFFLFFILNISNALFLVLDVKLFIPY*QKQIQEPLSSMRLCVI 263
VFS F+IFFL FI+N+S L L I + I++P +S+ L +
Sbjct: 235 VFSVFKIFFLLFIINVSIILLLKYSHNERIRMTSQSIRDPRTSIMLLAV 283
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 472,278,222
Number of Sequences: 1393205
Number of extensions: 9493973
Number of successful extensions: 29835
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 28414
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29756
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24568846532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)