Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003764A_C01 KMC003764A_c01
(858 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
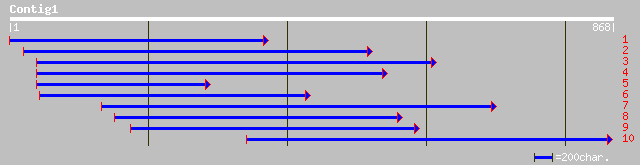
ref|NP_194093.1| putative protein; protein id: At4g23620.1, supp... 199 4e-50
dbj|BAB92303.1| P0451D05.2 [Oryza sativa (japonica cultivar-group)] 135 6e-31
ref|NP_201487.1| putative protein; protein id: At5g66860.1, supp... 108 8e-23
gb|ZP_00009136.1| hypothetical protein [Rhodopseudomonas palustris] 85 2e-15
ref|NP_103973.1| 50S ribosomal protein L25 [Mesorhizobium loti] ... 78 2e-13
>ref|NP_194093.1| putative protein; protein id: At4g23620.1, supported by cDNA:
6527., supported by cDNA: gi_18253004 [Arabidopsis
thaliana] gi|7486750|pir||T05594 hypothetical protein
F9D16.90 - Arabidopsis thaliana
gi|4454031|emb|CAA23028.1| putative protein [Arabidopsis
thaliana] gi|7269210|emb|CAB79317.1| putative protein
[Arabidopsis thaliana] gi|18253005|gb|AAL62429.1|
putative protein [Arabidopsis thaliana]
gi|21389687|gb|AAM48042.1| putative protein [Arabidopsis
thaliana] gi|21594031|gb|AAM65949.1| unknown
[Arabidopsis thaliana]
Length = 264
Score = 199 bits (506), Expect = 4e-50
Identities = 97/139 (69%), Positives = 117/139 (83%)
Frame = -1
Query: 855 LHVLSDFDSDEIIENVRVLPHQIHLQAGTDAPLNVTFLRAPSDALLRVDVPLVYRGDDVS 676
+ V ++ SDE+IE VR LP IHL +GTDAPLNVTF+RAP ALL+VD+PLV+ GDDVS
Sbjct: 121 VEVRAEIGSDEVIEKVRALPRAIHLHSGTDAPLNVTFIRAPPGALLKVDIPLVFIGDDVS 180
Query: 675 PGLKKGASLNTIRRTVKYLCPADIIPPYIDVDLSEMDVGQKLVMGDLIVHPALKLLHSKD 496
PGLKKGASLNTI+RTVK+LCPA+IIPPYI+VDLS++D+GQKLVMGDL VHPALKL+ SKD
Sbjct: 181 PGLKKGASLNTIKRTVKFLCPAEIIPPYIEVDLSQLDIGQKLVMGDLKVHPALKLIKSKD 240
Query: 495 EPVCKIMGPRAF*TTTTKE 439
EP+ K+ G R T T K+
Sbjct: 241 EPIVKVAGGRV--TDTQKK 257
>dbj|BAB92303.1| P0451D05.2 [Oryza sativa (japonica cultivar-group)]
Length = 393
Score = 135 bits (341), Expect = 6e-31
Identities = 61/83 (73%), Positives = 75/83 (89%)
Frame = -1
Query: 825 EIIENVRVLPHQIHLQAGTDAPLNVTFLRAPSDALLRVDVPLVYRGDDVSPGLKKGASLN 646
E+IE+VRVLP ++HL AGTD PLNVTF+RAPS ALL++DVPL++ G+D SPGL+KGA N
Sbjct: 144 ELIESVRVLPRKVHLHAGTDEPLNVTFMRAPSSALLKIDVPLMFIGEDASPGLRKGAYFN 203
Query: 645 TIRRTVKYLCPADIIPPYIDVDL 577
TI+RTVKYLCPADI+PPYI+VDL
Sbjct: 204 TIKRTVKYLCPADIVPPYIEVDL 226
>ref|NP_201487.1| putative protein; protein id: At5g66860.1, supported by cDNA:
gi_15450929, supported by cDNA: gi_17978762 [Arabidopsis
thaliana] gi|9758136|dbj|BAB08628.1|
gene_id:MUD21.12~pir||T05594~similar to unknown protein
[Arabidopsis thaliana] gi|15450930|gb|AAK96736.1|
Unknown protein [Arabidopsis thaliana]
gi|17978763|gb|AAL47375.1| unknown protein [Arabidopsis
thaliana]
Length = 249
Score = 108 bits (271), Expect = 8e-23
Identities = 59/130 (45%), Positives = 88/130 (67%), Gaps = 2/130 (1%)
Frame = -1
Query: 858 QLHVLSDFDSDEIIENVRVLPHQIHLQAGTDAPLNVTFLRAPSDALLRVDVPLVYRGDDV 679
QL + + S ++E+ RVLP ++H T LN+ F+ A L+VDVP+V++G D
Sbjct: 108 QLQIRAGQGSSTLVESGRVLPLKVHRDEETGKILNLVFVWADDGEKLKVDVPVVFKGLDH 167
Query: 678 SPGLKKGASLNTIRRTVKYLCPADIIPPYIDVDLSEMDVGQKLVMGDLIVHPALKLLHSK 499
PGL+KG +L TIR T+K L PA+ IP I+VD+S +D+ K+++ D++ HP+LKLL SK
Sbjct: 168 CPGLQKGGNLRTIRSTLKLLGPAEHIPSKIEVDVSNLDIEDKVLLQDVVFHPSLKLL-SK 226
Query: 498 DE--PVCKIM 475
+E PVCKI+
Sbjct: 227 NETMPVCKIV 236
>gb|ZP_00009136.1| hypothetical protein [Rhodopseudomonas palustris]
Length = 230
Score = 84.7 bits (208), Expect = 2e-15
Identities = 41/116 (35%), Positives = 68/116 (58%)
Frame = -1
Query: 807 RVLPHQIHLQAGTDAPLNVTFLRAPSDALLRVDVPLVYRGDDVSPGLKKGASLNTIRRTV 628
RV+P HL D P++V FLR + A +RV VPL +G +V+PG+K+G + N + TV
Sbjct: 74 RVIPRDYHLDPVRDFPIHVDFLRLGAGATIRVSVPLHLKGLEVAPGVKRGGTFNIVTHTV 133
Query: 627 KYLCPADIIPPYIDVDLSEMDVGQKLVMGDLIVHPALKLLHSKDEPVCKIMGPRAF 460
+ PA+ IP +I+ D+S +D+G L + D+ + +K + D + I+ P +
Sbjct: 134 ELEAPAENIPQFIEADVSTLDIGVSLHLSDIALPTGVKSVSRDDVTLVTIVPPSGY 189
>ref|NP_103973.1| 50S ribosomal protein L25 [Mesorhizobium loti]
gi|14023152|dbj|BAB49759.1| 50S ribosomal protein L25
[Mesorhizobium loti]
Length = 213
Score = 77.8 bits (190), Expect = 2e-13
Identities = 39/119 (32%), Positives = 68/119 (56%), Gaps = 1/119 (0%)
Frame = -1
Query: 816 ENVRVLPHQIHLQAGTDAPLNVTFLRAPSDALLRVDVPLVYRGDDVSPGLKKGASLNTIR 637
+ ++VLP L D P++V FLR D + VDVP+ + +D SPG+K+G LN +R
Sbjct: 72 KKIQVLPKDFQLDPVKDFPVHVDFLRIGKDTEVNVDVPVHFINEDKSPGIKRGGVLNIVR 131
Query: 636 RTVKYLCPADIIPPYIDVDLSEMDVGQKLVMGDLIVHPALK-LLHSKDEPVCKIMGPRA 463
V++ CPA+ IP +I +DL+ ++G + + + + +K ++ +D + I G A
Sbjct: 132 HEVEFHCPANAIPEFITIDLTGTNIGDSIHISAVQLPAGVKPVISDRDFTIATIAGSSA 190
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 758,171,016
Number of Sequences: 1393205
Number of extensions: 17240061
Number of successful extensions: 39076
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 37367
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39059
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 45431072631
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)