Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003761A_C01 KMC003761A_c01
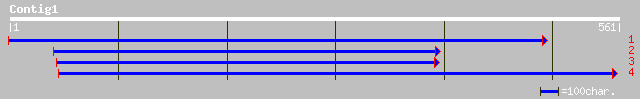
(558 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_566677.1| expressed protein; protein id: At3g21190.1, sup... 167 6e-41
gb|AAM65575.1| unknown [Arabidopsis thaliana] 167 6e-41
ref|NP_175574.1| unknown protein; protein id: At1g51630.1 [Arabi... 157 6e-38
gb|AAL84301.1|AC073556_18 putative auxin independent growth-rela... 45 6e-04
ref|NP_195552.1| putative growth regulator protein; protein id: ... 43 0.003
>ref|NP_566677.1| expressed protein; protein id: At3g21190.1, supported by cDNA:
40534. [Arabidopsis thaliana] gi|9280220|dbj|BAB01710.1|
emb|CAB10440.1~gene_id:MXL8.4~similar to unknown protein
[Arabidopsis thaliana] gi|28973699|gb|AAO64166.1|
unknown protein [Arabidopsis thaliana]
Length = 422
Score = 167 bits (424), Expect = 6e-41
Identities = 84/150 (56%), Positives = 108/150 (72%), Gaps = 5/150 (3%)
Frame = +1
Query: 124 MGVDLRQIVAGILTITMFVMLIHMIKRDNFDAVEDKLQGTTEEVIYENVKLDTTHE---- 291
MGVDLRQ+VAGILTITMFVML M+ RD FD++++K QG +++ +E K+
Sbjct: 1 MGVDLRQVVAGILTITMFVMLGQMLHRDYFDSLQEKAQGDAQDIEFEGSKVSVKDGLVGT 60
Query: 292 -RKNIGLGKGDADELKPCWAKPVGDDVERTEGFVIFSLTNGPEYHISQIADAVIVARSLG 468
+ GL D +L PCW + DD ++G+V FSLTNGPEYHISQI DAV+VA+ LG
Sbjct: 61 VEGSKGLWMEDNTDLTPCWPTLLSDDAVSSKGYVTFSLTNGPEYHISQITDAVMVAKHLG 120
Query: 469 ATLVIPDIRGSQPGEKRNFEDIYDVDVFMK 558
ATLV+PDIRGS+PG++RNFEDIYD D +K
Sbjct: 121 ATLVLPDIRGSKPGDERNFEDIYDADKLIK 150
>gb|AAM65575.1| unknown [Arabidopsis thaliana]
Length = 422
Score = 167 bits (424), Expect = 6e-41
Identities = 84/150 (56%), Positives = 108/150 (72%), Gaps = 5/150 (3%)
Frame = +1
Query: 124 MGVDLRQIVAGILTITMFVMLIHMIKRDNFDAVEDKLQGTTEEVIYENVKLDTTHE---- 291
MGVDLRQ+VAGILTITMFVML M+ RD FD++++K QG +++ +E K+
Sbjct: 1 MGVDLRQVVAGILTITMFVMLGQMLHRDYFDSLQEKAQGDAQDIEFEGSKVSVKDGLVGT 60
Query: 292 -RKNIGLGKGDADELKPCWAKPVGDDVERTEGFVIFSLTNGPEYHISQIADAVIVARSLG 468
+ GL D +L PCW + DD ++G+V FSLTNGPEYHISQI DAV+VA+ LG
Sbjct: 61 VEGSKGLWMEDNTDLTPCWPTLLSDDAVSSKGYVTFSLTNGPEYHISQITDAVMVAKHLG 120
Query: 469 ATLVIPDIRGSQPGEKRNFEDIYDVDVFMK 558
ATLV+PDIRGS+PG++RNFEDIYD D +K
Sbjct: 121 ATLVLPDIRGSKPGDERNFEDIYDADKLIK 150
>ref|NP_175574.1| unknown protein; protein id: At1g51630.1 [Arabidopsis thaliana]
gi|25405488|pir||A96555 unknown protein [imported] -
Arabidopsis thaliana
gi|12321689|gb|AAG50891.1|AC025294_29 unknown protein
[Arabidopsis thaliana]
Length = 460
Score = 157 bits (398), Expect = 6e-38
Identities = 85/162 (52%), Positives = 107/162 (65%), Gaps = 17/162 (10%)
Frame = +1
Query: 124 MGVDLRQIVAGILTITMFVMLIHMIKRDNFDAVE-----------DKLQGTTEEVIYENV 270
MGVDLRQ+VAGILTITMFVML M+ RD FDAV+ +K+QG ++ +
Sbjct: 1 MGVDLRQVVAGILTITMFVMLGQMLHRDYFDAVQVQTLERMIDSAEKVQGDAHDIEFHGS 60
Query: 271 K------LDTTHERKNIGLGKGDADELKPCWAKPVGDDVERTEGFVIFSLTNGPEYHISQ 432
K L E G D+ ELKPCW+ D+ ++G+V FSLTNGPEYH+SQ
Sbjct: 61 KVAVEDGLVRAFEAGTKGPWMEDSHELKPCWSISQSDEAVSSKGYVTFSLTNGPEYHVSQ 120
Query: 433 IADAVIVARSLGATLVIPDIRGSQPGEKRNFEDIYDVDVFMK 558
I DAV+VA+ LGATLV+PDIRGS+PG++ FEDIYDVD +K
Sbjct: 121 ITDAVMVAKHLGATLVLPDIRGSKPGDEMKFEDIYDVDKLIK 162
>gb|AAL84301.1|AC073556_18 putative auxin independent growth-related protein [Oryza sativa
(japonica cultivar-group)]
Length = 466
Score = 45.1 bits (105), Expect = 6e-04
Identities = 30/81 (37%), Positives = 45/81 (55%), Gaps = 1/81 (1%)
Frame = +1
Query: 316 GDADELKPCWAKPVGDDVERTEGFVIFSLTNGPEYHISQIADAVIVARSLGATLVIPDI- 492
GDA+ P A P V ++ G++ S G S+I D V VAR L T+V+P++
Sbjct: 40 GDAEPSPPPTALPPRR-VYKSNGYLKVSCNGGLNQMRSEICDMVAVARLLNLTMVVPELD 98
Query: 493 RGSQPGEKRNFEDIYDVDVFM 555
+ S ++ NFEDI+DV F+
Sbjct: 99 KRSFWADQSNFEDIFDVKHFI 119
>ref|NP_195552.1| putative growth regulator protein; protein id: At4g38390.1
[Arabidopsis thaliana] gi|7488119|pir||T05667 probable
growth regulator F22I13.160 - Arabidopsis thaliana
gi|4539347|emb|CAB37495.1| putative growth regulator
protein [Arabidopsis thaliana]
gi|7270823|emb|CAB80504.1| putative growth regulator
protein [Arabidopsis thaliana]
Length = 551
Score = 42.7 bits (99), Expect = 0.003
Identities = 25/68 (36%), Positives = 39/68 (56%), Gaps = 1/68 (1%)
Frame = +1
Query: 355 VGDDVERTEGFVIFSLTNGPEYHISQIADAVIVARSLGATLVIPDI-RGSQPGEKRNFED 531
V D +T +++ + + G + I DAV+ A L ATLV+P + + S + NFED
Sbjct: 116 VTDTRSQTNRYLLIATSGGLNQQRTGIIDAVVAAYILNATLVVPKLDQKSYWKDTSNFED 175
Query: 532 IYDVDVFM 555
I+DVD F+
Sbjct: 176 IFDVDWFI 183
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 469,533,909
Number of Sequences: 1393205
Number of extensions: 10143360
Number of successful extensions: 24612
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 23833
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24585
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)