Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003754A_C01 KMC003754A_c01
(599 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
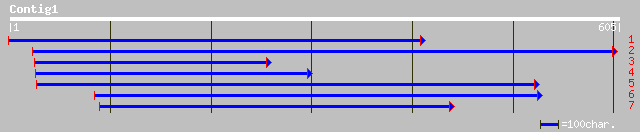
ref|NP_194639.1| putative protein; protein id: At4g29100.1, supp... 129 4e-29
emb|CAC14433.1| putative protein [Brassica napus] 111 8e-24
ref|NP_179600.1| unknown protein; protein id: At2g20100.1 [Arabi... 66 4e-10
gb|AAK63937.1|AC084282_18 putative ethylene-responsive protein [... 60 2e-08
gb|AAM62823.1| unknown [Arabidopsis thaliana] 59 6e-08
>ref|NP_194639.1| putative protein; protein id: At4g29100.1, supported by cDNA:
gi_19698938 [Arabidopsis thaliana]
gi|7485742|pir||T08965 hypothetical protein F19B15.130 -
Arabidopsis thaliana gi|4972056|emb|CAB43924.1| putative
protein [Arabidopsis thaliana]
gi|7269808|emb|CAB79668.1| putative protein [Arabidopsis
thaliana] gi|19698939|gb|AAL91205.1| putative protein
[Arabidopsis thaliana]
gi|22711852|gb|AAM10966.2|AF488634_1 putative bHLH
transcription factor [Arabidopsis thaliana]
gi|23197826|gb|AAN15440.1| putative protein [Arabidopsis
thaliana]
Length = 407
Score = 129 bits (323), Expect = 4e-29
Identities = 63/106 (59%), Positives = 76/106 (71%), Gaps = 8/106 (7%)
Frame = -2
Query: 598 FLQGQIEALSSPYLGN-GSKNMRNQQYVHGERNAVFPEDPGQLLNDSGLKRKGAPNQDGD 422
FLQ QIEALS PY G S NMR+QQ++ G+R+ +FPEDPGQL+ND +KR+GA + D
Sbjct: 301 FLQSQIEALSHPYFGTTASGNMRHQQHLQGDRSCIFPEDPGQLVNDQCMKRRGASSSSTD 360
Query: 421 RP-------KDLRSRGLCLVPVSCTQHVGNENGADYWAPGYGVGGF 305
KDLRSRGLCLVP+SCT VG++NGADYWAP G GF
Sbjct: 361 NQNASEEPKKDLRSRGLCLVPISCTLQVGSDNGADYWAPALGSAGF 406
>emb|CAC14433.1| putative protein [Brassica napus]
Length = 389
Score = 111 bits (277), Expect = 8e-24
Identities = 58/106 (54%), Positives = 71/106 (66%), Gaps = 8/106 (7%)
Frame = -2
Query: 598 FLQGQIEALSSPYLGN-GSKNMRNQQYVHGERNAVFPEDPGQLLNDSGLKRKGAPNQDGD 422
FLQ QIEALS PY G S NMRNQ ++ G+R+ +FPEDPGQL+ D +KR+G + +
Sbjct: 284 FLQNQIEALSHPYFGTTASGNMRNQ-HLQGDRSCLFPEDPGQLVTDQCVKRRGVSSSSSE 342
Query: 421 R-------PKDLRSRGLCLVPVSCTQHVGNENGADYWAPGYGVGGF 305
KDLRSRGLCLVP+S T VG++NGADYWAP G F
Sbjct: 343 NQNAKEEPKKDLRSRGLCLVPISYTLQVGSDNGADYWAPTLGSASF 388
>ref|NP_179600.1| unknown protein; protein id: At2g20100.1 [Arabidopsis thaliana]
gi|25411965|pir||A84585 hypothetical protein At2g20100
[imported] - Arabidopsis thaliana
gi|4580463|gb|AAD24387.1| unknown protein [Arabidopsis
thaliana]
Length = 59
Score = 65.9 bits (159), Expect = 4e-10
Identities = 29/45 (64%), Positives = 36/45 (79%)
Frame = -2
Query: 451 RKGAPNQDGDRPKDLRSRGLCLVPVSCTQHVGNENGADYWAPGYG 317
+K PN++ KDLRSRGLCLVP+SCT VG++NGADYWAP +G
Sbjct: 13 QKSNPNEEP--MKDLRSRGLCLVPISCTLQVGSDNGADYWAPAFG 55
>gb|AAK63937.1|AC084282_18 putative ethylene-responsive protein [Oryza sativa (japonica
cultivar-group)]
Length = 317
Score = 60.5 bits (145), Expect = 2e-08
Identities = 40/102 (39%), Positives = 49/102 (47%), Gaps = 7/102 (6%)
Frame = -2
Query: 598 FLQGQIEALSSPYLGNGSKNMRNQQYVHGERNAVFPEDPGQLLNDSGLKRKGAPNQDGDR 419
FL Q++ LSSPYL + R + G A E P L R
Sbjct: 197 FLHDQVQVLSSPYLQRLPPSARVPEQERGTPAA--EEQPPAL-----------------R 237
Query: 418 PKDLRSRGLCLVPVSCTQHV-------GNENGADYWAPGYGV 314
P DLRSRGLCLVP+SCT+HV G+ NGAD W+ G+
Sbjct: 238 PSDLRSRGLCLVPISCTEHVAGAGAGTGHGNGADLWSVAAGM 279
>gb|AAM62823.1| unknown [Arabidopsis thaliana]
Length = 393
Score = 58.5 bits (140), Expect = 6e-08
Identities = 34/98 (34%), Positives = 46/98 (46%)
Frame = -2
Query: 598 FLQGQIEALSSPYLGNGSKNMRNQQYVHGERNAVFPEDPGQLLNDSGLKRKGAPNQDGDR 419
FL Q+ LS+PY+ G+ N + QQ SG + +QD +
Sbjct: 318 FLHDQVTVLSTPYMKQGASNQQQQQI-------------------SGKSK----SQDENE 354
Query: 418 PKDLRSRGLCLVPVSCTQHVGNENGADYWAPGYGVGGF 305
+LR GLCLVP+S T V NE AD+W P +G F
Sbjct: 355 NHELRGHGLCLVPISSTFPVANETTADFWTPTFGGNNF 392
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 497,851,474
Number of Sequences: 1393205
Number of extensions: 10903367
Number of successful extensions: 21961
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 21294
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21945
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)