Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003749A_C01 KMC003749A_c01
(472 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
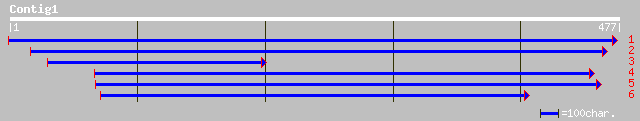
gb|EAA15613.1| SNF2 family N-terminal domain, putative [Plasmodi... 33 1.1
pir||T14555 DNA polymerase homolog dpo-r - beet mitochondrion gi... 33 2.0
ref|NP_064018.1| orf409 [Beta vulgaris] gi|9838500|ref|NP_064112... 33 2.0
ref|NP_703553.1| hypothetical protein [Plasmodium falciparum 3D7... 32 2.6
ref|NP_704307.1| hypothetical protein [Plasmodium falciparum 3D7... 32 3.3
>gb|EAA15613.1| SNF2 family N-terminal domain, putative [Plasmodium yoelii yoelii]
Length = 986
Score = 33.5 bits (75), Expect = 1.1
Identities = 20/53 (37%), Positives = 27/53 (50%)
Frame = -2
Query: 237 VLDSHISQSSRRTSVGKETKKEEKKMNDRNESSKC*SVKNIL*Q**GLNNDDD 79
VLDS S +S + ++ KE K E+KK ND N++ NNDDD
Sbjct: 55 VLDSSNSDNSDQENIKKECKNEKKKKNDNNDNDD------------NDNNDDD 95
>pir||T14555 DNA polymerase homolog dpo-r - beet mitochondrion
gi|1143422|emb|CAA84068.1| dpo-r [Beta vulgaris]
Length = 774
Score = 32.7 bits (73), Expect = 2.0
Identities = 15/32 (46%), Positives = 22/32 (67%)
Frame = -1
Query: 427 DSLYSFVKSFLCCPLSSLDGLILSSFLPFHKV 332
DSLY F+++++ CP S I+ FLP+HKV
Sbjct: 371 DSLYGFIEAYVECPES-----IVRPFLPYHKV 397
>ref|NP_064018.1| orf409 [Beta vulgaris] gi|9838500|ref|NP_064112.1| orf409 [Beta
vulgaris] gi|9049319|dbj|BAA99329.1| orf409 [Beta
vulgaris] gi|9087362|dbj|BAA99506.1| orf409 [Beta
vulgaris]
Length = 409
Score = 32.7 bits (73), Expect = 2.0
Identities = 15/32 (46%), Positives = 22/32 (67%)
Frame = -1
Query: 427 DSLYSFVKSFLCCPLSSLDGLILSSFLPFHKV 332
DSLY F+++++ CP S I+ FLP+HKV
Sbjct: 371 DSLYGFIEAYVECPES-----IVRPFLPYHKV 397
>ref|NP_703553.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23504695|emb|CAD51573.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 2054
Score = 32.3 bits (72), Expect = 2.6
Identities = 29/127 (22%), Positives = 51/127 (39%), Gaps = 8/127 (6%)
Frame = +3
Query: 75 EHHHHYSNLIIATKYSSQINI*NSHFYHSFSFPLFLFPSPPKFFLNFVRYENQE----PL 242
E+++ N + TKY +IN N H Y + + + F +++E PL
Sbjct: 208 EYYYDIQNECVNTKYYDKINQINDHIYKEEN----IMEDSSIQLMPFCNRKDKEKICQPL 263
Query: 243 HKYDFHSLAISATDSYITTRLQKAMPTYQPTLWNGRNELRINPSKEERGQ----HRKDLT 410
K+ L+ + T + K P N N + NP+++E H+K T
Sbjct: 264 TKHINEKLSYDFYTNNTTDNVNKKENHLSPKECNNNNNIFDNPNEKENSLKKPFHQKKTT 323
Query: 411 KLYKESV 431
L +E +
Sbjct: 324 TLNEEEI 330
>ref|NP_704307.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23499046|emb|CAD51126.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 643
Score = 32.0 bits (71), Expect = 3.3
Identities = 23/62 (37%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Frame = +1
Query: 10 FY*HIKNKRINIIYTNKCLYSMSIIIIIQTLSLLQNILHRLTFRTLISIIHFLFLFF--- 180
F+ ++ KRI I K + I+I + L+QN +H FR L S F+FLFF
Sbjct: 120 FWEYVIRKRIKI----KTFCKLGILISVFNF-LIQNNIHNDFFRILTSSFFFVFLFFLHI 174
Query: 181 CF 186
CF
Sbjct: 175 CF 176
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 385,766,469
Number of Sequences: 1393205
Number of extensions: 7775107
Number of successful extensions: 24366
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 23007
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24243
length of database: 448,689,247
effective HSP length: 113
effective length of database: 291,257,082
effective search space used: 12524054526
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)