Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003747A_C01 KMC003747A_c01
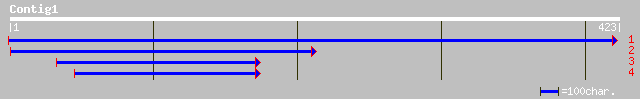
(420 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_704092.1| zinc transporter, putative [Plasmodium falcipar... 32 2.4
ref|ZP_00116367.1| hypothetical protein [Synechococcus sp. WH 8102] 32 2.4
ref|NP_495802.1| Putative cytoplasmic protein, nematode specific... 31 4.1
emb|CAA98065.2| Hypothetical protein ZK617.1b [Caenorhabditis el... 30 7.0
ref|NP_502274.1| UNCoordinated locomotion UNC-22, Immunoglobulin... 30 7.0
>ref|NP_704092.1| zinc transporter, putative [Plasmodium falciparum 3D7]
gi|23498830|emb|CAD50907.1| zinc transporter, putative
[Plasmodium falciparum 3D7]
Length = 556
Score = 31.6 bits (70), Expect = 2.4
Identities = 17/67 (25%), Positives = 29/67 (42%), Gaps = 9/67 (13%)
Frame = +3
Query: 24 YFSYVTEDPKTITFTE---------LGRGVYIEKNIYNSANKEL*TDSSRKRARGGGERD 176
Y+ + ED K + + + Y+ KN+YNS NK SS+ +A + +
Sbjct: 252 YYDNMEEDDKNVGHVNDMSEQNNNFMNKEKYMSKNVYNSTNKNSNNSSSKGKASNIQQNN 311
Query: 177 YNEKKNL 197
N N+
Sbjct: 312 LNNNNNI 318
>ref|ZP_00116367.1| hypothetical protein [Synechococcus sp. WH 8102]
Length = 661
Score = 31.6 bits (70), Expect = 2.4
Identities = 25/77 (32%), Positives = 36/77 (46%), Gaps = 1/77 (1%)
Frame = +3
Query: 66 TELGRGVYIEKNIYNSANKEL*TDSS-RKRARGGGERDYNEKKNLFACAYLTSLFKKNSA 242
TE+ +G +EK+ + +++ S +K A GGG R YN + F Y L K S
Sbjct: 234 TEIIKGTKLEKHYKDIRSRKFRDHKSWKKDATGGGTRAYNSQYGAFTFNYEQKLNLKTSF 293
Query: 243 KCK*LRKCRRNIYTSFT 293
K L +Y SFT
Sbjct: 294 TGKDL------LYASFT 304
>ref|NP_495802.1| Putative cytoplasmic protein, nematode specific [Caenorhabditis
elegans] gi|7508766|pir||T26049 hypothetical protein
W01C9.1 - Caenorhabditis elegans
gi|3880378|emb|CAA90266.1| Hypothetical protein W01C9.1
[Caenorhabditis elegans]
Length = 258
Score = 30.8 bits (68), Expect = 4.1
Identities = 18/67 (26%), Positives = 32/67 (46%)
Frame = -1
Query: 213 NRHKRIGFSFHYNLSPPPPALSFSMSLFTILYLLNCIYFFQYIPPFPIQ*M**FLGLQLH 34
++H F FH+++ P A + +I +L Y++++ P + F+G Q
Sbjct: 30 HKHSTRSFHFHFDVFAPYSAFTLRYLGISIFHLHYNNYYYKFYYPMLFLILTVFVGAQAA 89
Query: 33 KKSMEDV 13
KK M DV
Sbjct: 90 KKPMFDV 96
>emb|CAA98065.2| Hypothetical protein ZK617.1b [Caenorhabditis elegans]
gi|26985881|emb|CAA98082.2| Hypothetical protein
ZK617.1b [Caenorhabditis elegans]
Length = 7158
Score = 30.0 bits (66), Expect = 7.0
Identities = 13/36 (36%), Positives = 21/36 (58%)
Frame = -1
Query: 405 VRKTESVDSPGSRTSDTPSQAAVLCNETEEASIDEP 298
++K E DS GS+ D P + ++ T ++S DEP
Sbjct: 795 LQKYEKTDSDGSKKEDKPKKVSIAPVSTNKSSDDEP 830
>ref|NP_502274.1| UNCoordinated locomotion UNC-22, Immunoglobulin and fibronectin
type III domains containing protein kinase family member
(unc-22) [Caenorhabditis elegans] gi|7511244|pir||T27935
hypothetical protein ZK617.1b - Caenorhabditis elegans
Length = 7160
Score = 30.0 bits (66), Expect = 7.0
Identities = 13/36 (36%), Positives = 21/36 (58%)
Frame = -1
Query: 405 VRKTESVDSPGSRTSDTPSQAAVLCNETEEASIDEP 298
++K E DS GS+ D P + ++ T ++S DEP
Sbjct: 797 LQKYEKTDSDGSKKEDKPKKVSIAPVSTNKSSDDEP 832
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 355,311,698
Number of Sequences: 1393205
Number of extensions: 7305885
Number of successful extensions: 16623
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 16115
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 16576
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 6923296128
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)