Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003743A_C01 KMC003743A_c01
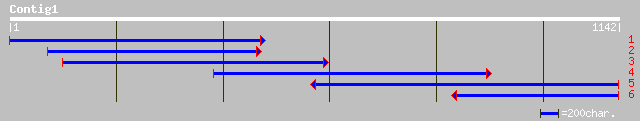
(1140 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T08942 proton pump interactor - Arabidopsis thaliana gi|497... 265 1e-69
ref|NP_194480.2| proton pump interactor; protein id: At4g27500.1... 265 1e-69
ref|NP_568541.1| putative protein; protein id: At5g36780.1 [Arab... 170 4e-41
dbj|BAB02163.1| gb|AAF34843.1~gene_id:K7L4.14~strong similarity ... 169 1e-40
dbj|BAA97551.1| contains similarity to proton pump interactor~ge... 132 1e-29
>pir||T08942 proton pump interactor - Arabidopsis thaliana
gi|4972075|emb|CAB43882.1| proton pump interactor
[Arabidopsis thaliana] gi|7269604|emb|CAB81400.1| proton
pump interactor [Arabidopsis thaliana]
Length = 628
Score = 265 bits (677), Expect = 1e-69
Identities = 153/292 (52%), Positives = 196/292 (66%), Gaps = 12/292 (4%)
Frame = +2
Query: 8 RELLRKKDINAIDELSQTEVEKFMSLWNSDKSFRNDYEKRMLQSLDMRQLSRDGRMRNPD 187
R+L +K+I+ ++ L+ EVEKF+SLW S K+FR DYEKR+LQSLD RQLSRDGRMRNPD
Sbjct: 341 RDLAAQKNISELEALANAEVEKFISLWCSKKNFREDYEKRILQSLDSRQLSRDGRMRNPD 400
Query: 188 EKPLL------EEPKPAETDAFSKAIPK-QPKEEPKPSPQETLPTQKESKNKVRDLKAKP 346
EKPL+ + P+ET+ KA K QPKEEP +P+ T ++ K +D
Sbjct: 401 EKPLIAPEAAPSKATPSETEVVPKAKAKPQPKEEPVSAPKPDA-TVAQNTEKAKDAVKVK 459
Query: 347 DKKDLEEADEYEFEVPRKETPPKEPEIDPAKLKEMKREEEIAKAKQAWERKKKLAEKAAA 526
+ D ++ + Y P+KE P +D A KEM+++EEIAKAKQA ERKKKLAEKAAA
Sbjct: 460 NVADDDDDEVYGLGKPQKEEKP----VDAATAKEMRKQEEIAKAKQAMERKKKLAEKAAA 515
Query: 527 KAALKAQKEAEKKLKK-----AKKKAGIVEEPADEVVEAPQEETINDNVEAAAPVKEKVP 691
KAA++AQKEAEKK KK AKKK G E E V EE I V+ P KEKV
Sbjct: 516 KAAIRAQKEAEKKEKKEQEKKAKKKTGGNTETETEEVPEASEEEIEAPVQEEKPQKEKVF 575
Query: 692 KESVIRSRSRAKGPDSLPKGILKRKRSNNYWIWIASAALLVLLLSVLGYIYL 847
KE IR+R+R +GP+++P+ ILKRK+S NYW++ A AAL+VLLL VLGY Y+
Sbjct: 576 KEKPIRNRTRGRGPETIPRAILKRKKSTNYWVYAAPAALVVLLLLVLGYYYV 627
>ref|NP_194480.2| proton pump interactor; protein id: At4g27500.1 [Arabidopsis
thaliana] gi|13992437|emb|CAA05145.2| proton pump
interactor [Arabidopsis thaliana]
gi|15215674|gb|AAK91382.1| AT4g27500/F27G19_100
[Arabidopsis thaliana] gi|23505987|gb|AAN28853.1|
At4g27500/F27G19_100 [Arabidopsis thaliana]
Length = 612
Score = 265 bits (677), Expect = 1e-69
Identities = 153/292 (52%), Positives = 196/292 (66%), Gaps = 12/292 (4%)
Frame = +2
Query: 8 RELLRKKDINAIDELSQTEVEKFMSLWNSDKSFRNDYEKRMLQSLDMRQLSRDGRMRNPD 187
R+L +K+I+ ++ L+ EVEKF+SLW S K+FR DYEKR+LQSLD RQLSRDGRMRNPD
Sbjct: 325 RDLAAQKNISELEALANAEVEKFISLWCSKKNFREDYEKRILQSLDSRQLSRDGRMRNPD 384
Query: 188 EKPLL------EEPKPAETDAFSKAIPK-QPKEEPKPSPQETLPTQKESKNKVRDLKAKP 346
EKPL+ + P+ET+ KA K QPKEEP +P+ T ++ K +D
Sbjct: 385 EKPLIAPEAAPSKATPSETEVVPKAKAKPQPKEEPVSAPKPDA-TVAQNTEKAKDAVKVK 443
Query: 347 DKKDLEEADEYEFEVPRKETPPKEPEIDPAKLKEMKREEEIAKAKQAWERKKKLAEKAAA 526
+ D ++ + Y P+KE P +D A KEM+++EEIAKAKQA ERKKKLAEKAAA
Sbjct: 444 NVADDDDDEVYGLGKPQKEEKP----VDAATAKEMRKQEEIAKAKQAMERKKKLAEKAAA 499
Query: 527 KAALKAQKEAEKKLKK-----AKKKAGIVEEPADEVVEAPQEETINDNVEAAAPVKEKVP 691
KAA++AQKEAEKK KK AKKK G E E V EE I V+ P KEKV
Sbjct: 500 KAAIRAQKEAEKKEKKEQEKKAKKKTGGNTETETEEVPEASEEEIEAPVQEEKPQKEKVF 559
Query: 692 KESVIRSRSRAKGPDSLPKGILKRKRSNNYWIWIASAALLVLLLSVLGYIYL 847
KE IR+R+R +GP+++P+ ILKRK+S NYW++ A AAL+VLLL VLGY Y+
Sbjct: 560 KEKPIRNRTRGRGPETIPRAILKRKKSTNYWVYAAPAALVVLLLLVLGYYYV 611
>ref|NP_568541.1| putative protein; protein id: At5g36780.1 [Arabidopsis thaliana]
gi|22327405|ref|NP_198484.2| putative protein; protein
id: At5g36690.1 [Arabidopsis thaliana]
Length = 576
Score = 170 bits (430), Expect = 4e-41
Identities = 111/300 (37%), Positives = 164/300 (54%), Gaps = 25/300 (8%)
Frame = +2
Query: 8 RELLRKKDINAIDELSQTEVEKFMSLWNSDKSFRNDYEKRMLQSLDMRQLSRDGRMRNPD 187
+EL ++ ++ + +EV++FM+LWN DK+FR DY +R+ SL R+L+ DGR+++ D
Sbjct: 294 KELAASGNVRELEVFASSEVDRFMTLWNDDKAFREDYVRRISHSLCERELNEDGRIKDAD 353
Query: 188 EKPLLEEPKPAETDAFSKAIPKQPKEEPKPSPQETLPTQKESKNKVRDLKAKPDKKDLEE 367
+ E+ P +T S+ + K +E+ + E N + D + K K D+
Sbjct: 354 LQIFWEKKVPVKTIKRSEKVHKMNREDSSSN-------SSEDGNVITDKRKKETKSDV-- 404
Query: 368 ADEYEFEVPRKETPPKEPEIDPAKLKEMKREEEIAKAKQAWERKKKLAEKAAAKAALKAQ 547
+E P+K KE EID LKE KREE++ KA+ ERK+KL EKAAAKAA++AQ
Sbjct: 405 ---IVYEKPKK----KEEEIDEEALKERKREEQLEKARLVMERKRKLQEKAAAKAAIRAQ 457
Query: 548 KEAEKKL---------------KKAKKKAGIVEEPADEVVEAPQEETINDNVEAAAPV-- 676
KEAEKKL KKAKKKA A+ + ++ IND V
Sbjct: 458 KEAEKKLKAIILSCSHFFNECEKKAKKKAA-----ANSTSPSESDQVINDEKVRTLAVSG 512
Query: 677 KEK-------VPKESVIRSRSRAKGPDSLPKGILKRKRSNNYWIW-IASAALLVLLLSVL 832
KEK PK+ R + R G ++LPK IL R+++ YW+W ++SAAL V L+ V+
Sbjct: 513 KEKHQKERLLFPKQRSFRYKHRGSGTEALPKAILNRRKARRYWVWGLSSAALAVSLVLVV 572
>dbj|BAB02163.1| gb|AAF34843.1~gene_id:K7L4.14~strong similarity to unknown protein
[Arabidopsis thaliana]
Length = 620
Score = 169 bits (427), Expect = 1e-40
Identities = 106/294 (36%), Positives = 171/294 (58%), Gaps = 19/294 (6%)
Frame = +2
Query: 8 RELLRKKDINAIDELSQTEVEKFMSLWNSDKSFRNDYEKRMLQSLDMRQLSRDGRMRNPD 187
REL ++ ++ + +E ++FM+ WN DK+FR+DY KR+ SL RQL++DGR+++P+
Sbjct: 330 RELAASGNVRDLEVFASSEADRFMTQWNDDKAFRDDYVKRISPSLCERQLNQDGRIKDPE 389
Query: 188 EKPLLEEPKPAETDAFSKAIPKQPKEEPKPSPQE-----TLPTQKESKNKVRDL-KAKPD 349
+ + E+ P + + + + +E+ + + T +KE++ K D ++ +
Sbjct: 390 VQVVWEKKVPVKG---GEKVHETNREDSSSNSSQYGSVITDKRKKETRKKAMDFNRSSAE 446
Query: 350 KKDLEEADEYEFEVPRKETPPKEPEIDPAKLKEMKREEEIAKAKQAWERKKKLAEKAAAK 529
+ D+ + + +E P+KE E E+D LKE +REE++ KA+ A ERK+KL EKAAAK
Sbjct: 447 ESDVTDLEFSVYEKPKKE----EEEVDEETLKEREREEQLEKARLAMERKRKLQEKAAAK 502
Query: 530 AALKAQKEAEKKL----KKAKKKAGIVEEPADEVVEAPQEETINDNVEA-AAPVKEK--- 685
AA++AQKEAEKKL KKAKKKA E + + + V A KEK
Sbjct: 503 AAIRAQKEAEKKLKECEKKAKKKAAANSSSPSESDHSQEVTKDLEKVRTLAVSGKEKHQK 562
Query: 686 ----VPKESVIRSRSRAKGPDSLPKGILKRKRSNNYWIW-IASAALLVLLLSVL 832
PK+ R + R +G ++LPK IL R++++ YW+W ++SAAL V L V+
Sbjct: 563 ERSLFPKQRSFRYKHRGRGTEALPKAILNRRKAHKYWVWGLSSAALAVALFLVV 616
>dbj|BAA97551.1| contains similarity to proton pump interactor~gene_id:F24C7.7
[Arabidopsis thaliana] gi|8953698|dbj|BAA98056.1|
contains similarity to proton pump
interactor~gene_id:F5H8.2 [Arabidopsis thaliana]
Length = 503
Score = 132 bits (332), Expect = 1e-29
Identities = 76/188 (40%), Positives = 113/188 (59%)
Frame = +2
Query: 8 RELLRKKDINAIDELSQTEVEKFMSLWNSDKSFRNDYEKRMLQSLDMRQLSRDGRMRNPD 187
+EL ++ ++ + +EV++FM+LWN DK+FR DY +R+ SL R+L+ DGR+++ D
Sbjct: 285 KELAASGNVRELEVFASSEVDRFMTLWNDDKAFREDYVRRISHSLCERELNEDGRIKDAD 344
Query: 188 EKPLLEEPKPAETDAFSKAIPKQPKEEPKPSPQETLPTQKESKNKVRDLKAKPDKKDLEE 367
+ E+ P +T S+ + K +E+ + E N + D + K K D+
Sbjct: 345 LQIFWEKKVPVKTIKRSEKVHKMNREDSSSN-------SSEDGNVITDKRKKETKSDV-- 395
Query: 368 ADEYEFEVPRKETPPKEPEIDPAKLKEMKREEEIAKAKQAWERKKKLAEKAAAKAALKAQ 547
+E P+K KE EID LKE KREE++ KA+ ERK+KL EKAAAKAA++AQ
Sbjct: 396 ---IVYEKPKK----KEEEIDEEALKERKREEQLEKARLVMERKRKLQEKAAAKAAIRAQ 448
Query: 548 KEAEKKLK 571
KEAEKKLK
Sbjct: 449 KEAEKKLK 456
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 976,469,205
Number of Sequences: 1393205
Number of extensions: 22883602
Number of successful extensions: 150766
Number of sequences better than 10.0: 4708
Number of HSP's better than 10.0 without gapping: 101755
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 130645
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 69732809988
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)