Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003742A_C01 KMC003742A_c01
(547 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
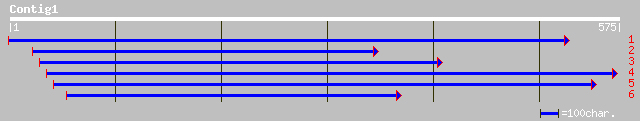
emb|CAA98164.1| RAB1Y [Lotus japonicus] 154 9e-37
pir||A96503 protein F9C16.3 [imported] - Arabidopsis thaliana gi... 120 8e-27
ref|NP_175056.1| GTP-binding protein(RAB1Y), putative; protein i... 120 8e-27
emb|CAA98163.1| RAB1X [Lotus japonicus] 96 2e-19
ref|NP_568121.1| putative protein; protein id: At5g03530.1, supp... 95 6e-19
>emb|CAA98164.1| RAB1Y [Lotus japonicus]
Length = 207
Score = 154 bits (388), Expect = 9e-37
Identities = 75/76 (98%), Positives = 75/76 (98%)
Frame = -3
Query: 545 DRXVSKKEGIDFAREYGCLFIECSAKTRVNVQQCFEELVLKILDTPSLLAEGSKGVKKNI 366
DR VSKKEGIDFAREYGCLFIECSAKTRVNVQQCFEELVLKILDTPSLLAEGSKGVKKNI
Sbjct: 132 DRVVSKKEGIDFAREYGCLFIECSAKTRVNVQQCFEELVLKILDTPSLLAEGSKGVKKNI 191
Query: 365 FKDKQPQADASTSSCC 318
FKDKQPQADASTSSCC
Sbjct: 192 FKDKQPQADASTSSCC 207
>pir||A96503 protein F9C16.3 [imported] - Arabidopsis thaliana
gi|8778652|gb|AAF79660.1|AC022314_1 F9C16.3 [Arabidopsis
thaliana]
Length = 233
Score = 120 bits (302), Expect = 8e-27
Identities = 58/76 (76%), Positives = 65/76 (85%)
Frame = -3
Query: 545 DRXVSKKEGIDFAREYGCLFIECSAKTRVNVQQCFEELVLKILDTPSLLAEGSKGVKKNI 366
+R VSKKEGIDFAREYGCLF+ECSAKTRVNV+QCFEELVLKIL+TPSL AEGS G KKNI
Sbjct: 155 ERAVSKKEGIDFAREYGCLFLECSAKTRVNVEQCFEELVLKILETPSLTAEGSSGGKKNI 214
Query: 365 FKDKQPQADASTSSCC 318
FK Q +++SS C
Sbjct: 215 FKQNPAQTTSTSSSYC 230
>ref|NP_175056.1| GTP-binding protein(RAB1Y), putative; protein id: At1g43890.1,
supported by cDNA: 2898., supported by cDNA:
gi_17104730, supported by cDNA: gi_2231311 [Arabidopsis
thaliana] gi|2231312|gb|AAB61997.1| AtRab18 [Arabidopsis
thaliana] gi|7523663|gb|AAF63103.1|AC006423_4 AtRab18
[Arabidopsis thaliana]
gi|13878001|gb|AAK44078.1|AF370263_1 putative
GTP-binding protein RAB1Y [Arabidopsis thaliana]
gi|17104731|gb|AAL34254.1| putative GTP-binding protein
RAB1Y [Arabidopsis thaliana] gi|21555736|gb|AAM63924.1|
GTP-binding protein(RAB1Y), putative [Arabidopsis
thaliana]
Length = 212
Score = 120 bits (302), Expect = 8e-27
Identities = 58/76 (76%), Positives = 65/76 (85%)
Frame = -3
Query: 545 DRXVSKKEGIDFAREYGCLFIECSAKTRVNVQQCFEELVLKILDTPSLLAEGSKGVKKNI 366
+R VSKKEGIDFAREYGCLF+ECSAKTRVNV+QCFEELVLKIL+TPSL AEGS G KKNI
Sbjct: 134 ERAVSKKEGIDFAREYGCLFLECSAKTRVNVEQCFEELVLKILETPSLTAEGSSGGKKNI 193
Query: 365 FKDKQPQADASTSSCC 318
FK Q +++SS C
Sbjct: 194 FKQNPAQTTSTSSSYC 209
>emb|CAA98163.1| RAB1X [Lotus japonicus]
Length = 208
Score = 96.3 bits (238), Expect = 2e-19
Identities = 45/75 (60%), Positives = 54/75 (72%)
Frame = -3
Query: 542 RXVSKKEGIDFAREYGCLFIECSAKTRVNVQQCFEELVLKILDTPSLLAEGSKGVKKNIF 363
R VS++EG+ A+E GCL IECSAKTR NV+QCFEEL LKI++ PSLL EGS VK+NI
Sbjct: 133 RAVSREEGLALAKELGCLLIECSAKTRENVEQCFEELALKIMEAPSLLEEGSTAVKRNIL 192
Query: 362 KDKQPQADASTSSCC 318
K KQ + CC
Sbjct: 193 KQKQEPQASQHGGCC 207
>ref|NP_568121.1| putative protein; protein id: At5g03530.1, supported by cDNA:
gi_2723476 [Arabidopsis thaliana]
gi|11274537|pir||T48379 AtRab GTP-binding protein -
Arabidopsis thaliana gi|2723477|dbj|BAA24074.1|
GTP-binding protein [Arabidopsis thaliana]
gi|7378638|emb|CAB83314.1| AtRab GTP-binding protein
[Arabidopsis thaliana] gi|26453054|dbj|BAC43603.1|
putative AtRab GTP-binding protein [Arabidopsis
thaliana]
Length = 210
Score = 94.7 bits (234), Expect = 6e-19
Identities = 43/76 (56%), Positives = 55/76 (71%)
Frame = -3
Query: 545 DRXVSKKEGIDFAREYGCLFIECSAKTRVNVQQCFEELVLKILDTPSLLAEGSKGVKKNI 366
+R VS++EGI A+E C+F+ECSA+TR NV+QCFEEL LKI++ PSLL EGS VK+NI
Sbjct: 134 ERGVSREEGIALAKELNCMFLECSARTRQNVEQCFEELALKIMEVPSLLEEGSSAVKRNI 193
Query: 365 FKDKQPQADASTSSCC 318
K K + S CC
Sbjct: 194 LKQKPEHQTNTQSGCC 209
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 423,623,200
Number of Sequences: 1393205
Number of extensions: 7914432
Number of successful extensions: 17489
Number of sequences better than 10.0: 676
Number of HSP's better than 10.0 without gapping: 17114
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 17486
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18660035355
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)