Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
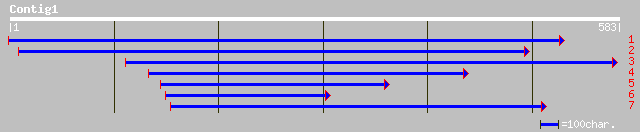
Query= KMC003739A_C01 KMC003739A_c01
(583 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_181891.2| unknown protein; protein id: At2g43630.1, suppo... 70 3e-11
pir||E84868 hypothetical protein At2g43630 [imported] - Arabidop... 70 3e-11
ref|NP_191523.1| putative protein; protein id: At3g59640.1, supp... 66 3e-10
emb|CAD41536.1| OSJNBb0091E11.5 [Oryza sativa (japonica cultivar... 64 1e-09
ref|NP_370781.1| hypothetical protein [Staphylococcus aureus sub... 36 0.31
>ref|NP_181891.2| unknown protein; protein id: At2g43630.1, supported by cDNA:
gi_17529329, supported by cDNA: gi_20465906 [Arabidopsis
thaliana] gi|17529330|gb|AAL38892.1| unknown protein
[Arabidopsis thaliana] gi|20465907|gb|AAM20106.1|
unknown protein [Arabidopsis thaliana]
Length = 274
Score = 69.7 bits (169), Expect = 3e-11
Identities = 38/106 (35%), Positives = 66/106 (61%), Gaps = 4/106 (3%)
Frame = -1
Query: 580 AGLSDEIQQIILATIGFILVYICVIDGVELAKLSKDILKYLFGGTQSLRLKKAIYKWTRF 401
AG++DE Q++LAT+GFI +Y +I G EL KL++D +++L G +++RL +A+ W F
Sbjct: 157 AGIADETLQVVLATLGFIFLYTYIITGEELVKLARDYIRFLMGRPKTVRLTRAMDSWNGF 216
Query: 400 FKLLTGNRKVLKNELEEA----PTQRNNLDYYHDDVLRNYMKPNSD 275
+ ++ R + LE+A PT ++ + Y V++ Y+ NSD
Sbjct: 217 LEKMSRQRVYDEYWLEKAIINTPTWYDSPEKYR-RVIKAYVDSNSD 261
>pir||E84868 hypothetical protein At2g43630 [imported] - Arabidopsis thaliana
gi|2281107|gb|AAB64043.1| unknown protein [Arabidopsis
thaliana]
Length = 174
Score = 69.7 bits (169), Expect = 3e-11
Identities = 38/106 (35%), Positives = 66/106 (61%), Gaps = 4/106 (3%)
Frame = -1
Query: 580 AGLSDEIQQIILATIGFILVYICVIDGVELAKLSKDILKYLFGGTQSLRLKKAIYKWTRF 401
AG++DE Q++LAT+GFI +Y +I G EL KL++D +++L G +++RL +A+ W F
Sbjct: 57 AGIADETLQVVLATLGFIFLYTYIITGEELVKLARDYIRFLMGRPKTVRLTRAMDSWNGF 116
Query: 400 FKLLTGNRKVLKNELEEA----PTQRNNLDYYHDDVLRNYMKPNSD 275
+ ++ R + LE+A PT ++ + Y V++ Y+ NSD
Sbjct: 117 LEKMSRQRVYDEYWLEKAIINTPTWYDSPEKYR-RVIKAYVDSNSD 161
>ref|NP_191523.1| putative protein; protein id: At3g59640.1, supported by cDNA:
gi_14423405 [Arabidopsis thaliana]
gi|11358054|pir||T49305 hypothetical protein T16L24.190
- Arabidopsis thaliana gi|6996300|emb|CAB75461.1|
putative protein [Arabidopsis thaliana]
gi|14423406|gb|AAK62385.1|AF386940_1 putative protein
[Arabidopsis thaliana] gi|24899691|gb|AAN65060.1|
putative protein [Arabidopsis thaliana]
Length = 246
Score = 66.2 bits (160), Expect = 3e-10
Identities = 31/74 (41%), Positives = 51/74 (68%)
Frame = -1
Query: 580 AGLSDEIQQIILATIGFILVYICVIDGVELAKLSKDILKYLFGGTQSLRLKKAIYKWTRF 401
A DE Q++LAT+GFI +Y +I+G EL +L++D ++YL G +S+RL + + W+RF
Sbjct: 151 ASFGDETLQVVLATLGFIFLYFYIINGEELFRLARDYIRYLIGRPKSVRLTRVMEGWSRF 210
Query: 400 FKLLTGNRKVLKNE 359
F+ + +RK + NE
Sbjct: 211 FEKM--SRKKVYNE 222
>emb|CAD41536.1| OSJNBb0091E11.5 [Oryza sativa (japonica cultivar-group)]
Length = 337
Score = 64.3 bits (155), Expect = 1e-09
Identities = 34/102 (33%), Positives = 62/102 (60%), Gaps = 6/102 (5%)
Frame = -1
Query: 565 EIQQIILATIGFILVYICVIDGVELAKLSKDILKYLFGGTQSLRLKKAIYKWTRFFKLLT 386
E+ Q++LATI FIL+YI +I G EL +L++D +YL G ++ RLK+A+ W F + +T
Sbjct: 233 EMVQVLLATIAFILMYIHIIRGEELYRLARDYTRYLVTGKRTSRLKRAMLNWHNFCEGIT 292
Query: 385 GNRKVLKNELEEAPTQ----RNNLDYYH--DDVLRNYMKPNS 278
V ++ E + ++ + L + H +++ R Y +P++
Sbjct: 293 NKDSVQESTFERSTSEPMWWQQPLKFVHRIEELYRGYFRPHA 334
>ref|NP_370781.1| hypothetical protein [Staphylococcus aureus subsp. aureus Mu50]
gi|15925960|ref|NP_373493.1| hypothetical protein,
similar to teichoic acid biosynthesis protein B
[Staphylococcus aureus subsp. aureus N315]
gi|21281962|ref|NP_645048.1| ORFID:MW0233~hypothetical
protein, similar to teichoic acid biosynthesis protein B
[Staphylococcus aureus subsp. aureus MW2]
gi|25506683|pir||D89789 hypothetical protein SA0247
[imported] - Staphylococcus aureus (strain N315)
gi|13700173|dbj|BAB41471.1| SA0247 [Staphylococcus
aureus subsp. aureus N315] gi|14246024|dbj|BAB56419.1|
hypothetical protein [Staphylococcus aureus subsp.
aureus Mu50] gi|21203397|dbj|BAB94098.1| MW0233
[Staphylococcus aureus subsp. aureus MW2]
Length = 562
Score = 36.2 bits (82), Expect = 0.31
Identities = 24/107 (22%), Positives = 52/107 (48%), Gaps = 2/107 (1%)
Frame = -1
Query: 574 LSDEIQQIILATIGFILVYICVIDGVELAKLSKDILKYLFGGTQSLR--LKKAIYKWTRF 401
+S ++ + +L ++V + + + + K++ KY + + R + KAI+ T+F
Sbjct: 149 ISSDVNEFVLD----VVVTTPEVKSIYIVRKYKELRKYFRKQSFNTRQFIFKAIFNTTKF 204
Query: 400 FKLLTGNRKVLKNELEEAPTQRNNLDYYHDDVLRNYMKPNSDA*VLF 260
F L GN + + + PT N +Y ++++LR + D +F
Sbjct: 205 FHLKKGNTVLFTS--DSRPTMSGNFEYIYNEMLRQNLDKKYDIHTVF 249
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 462,081,615
Number of Sequences: 1393205
Number of extensions: 9658336
Number of successful extensions: 25244
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 24518
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25223
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)