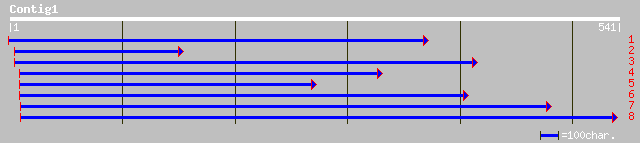
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003730A_C01 KMC003730A_c01
(534 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK84424.1|AF396465_1 peroxidase-like protein [Pisum sativum] 141 6e-33
gb|AAD11482.1| peroxidase precursor [Glycine max] 120 1e-26
gb|AAD11481.1| peroxidase precursor [Glycine max] 119 2e-26
gb|AAA65637.1| peroxidase 106 2e-22
pir||T09218 peroxidase (EC 1.11.1.7) precursor prx10 - spinach ... 105 4e-22
>gb|AAK84424.1|AF396465_1 peroxidase-like protein [Pisum sativum]
Length = 89
Score = 141 bits (355), Expect = 6e-33
Identities = 68/86 (79%), Positives = 76/86 (88%)
Frame = -1
Query: 531 TLIEMDPGSRNTFDLGYYKQVLKRRGLFESDAALLNSSVTRSIITQQLRSTQKFFLGFAK 352
TLIEMDPGSRNTFD+GY+KQV+KRRGLFESDAALL SS TRSI+ Q L+S +KFF FAK
Sbjct: 4 TLIEMDPGSRNTFDVGYFKQVVKRRGLFESDAALLKSSTTRSIVAQHLQSNEKFFTEFAK 63
Query: 351 SMEKMGRINVLTGTKGEIRKHCALVN 274
SMEKMGRINV GT+GEIRKHCA +N
Sbjct: 64 SMEKMGRINVKIGTEGEIRKHCAFIN 89
>gb|AAD11482.1| peroxidase precursor [Glycine max]
Length = 351
Score = 120 bits (300), Expect = 1e-26
Identities = 61/88 (69%), Positives = 70/88 (79%), Gaps = 1/88 (1%)
Frame = -1
Query: 531 TLIEMDPGSRNTFDLGYYKQVLKRRGLFESDAALLNSSVTRSIITQQLR-STQKFFLGFA 355
T IEMDPGSR TFDL YY V+KRRGLFESDAALL +SVT++ I Q L S + FF FA
Sbjct: 264 TKIEMDPGSRKTFDLSYYSHVIKRRGLFESDAALLTNSVTKAQIIQLLEGSVENFFAEFA 323
Query: 354 KSMEKMGRINVLTGTKGEIRKHCALVNN 271
S+EKMGRINV TGT+GEIRKHCA +N+
Sbjct: 324 TSIEKMGRINVKTGTEGEIRKHCAFINS 351
>gb|AAD11481.1| peroxidase precursor [Glycine max]
Length = 352
Score = 119 bits (299), Expect = 2e-26
Identities = 61/88 (69%), Positives = 70/88 (79%), Gaps = 1/88 (1%)
Frame = -1
Query: 531 TLIEMDPGSRNTFDLGYYKQVLKRRGLFESDAALLNSSVTRSIITQQLR-STQKFFLGFA 355
T IEMDPGSR TFDL YY V+KRRGLFESDAALL +SVT++ I + L S + FF FA
Sbjct: 265 TKIEMDPGSRKTFDLSYYSHVIKRRGLFESDAALLTNSVTKAQIIELLEGSVENFFAEFA 324
Query: 354 KSMEKMGRINVLTGTKGEIRKHCALVNN 271
SMEKMGRINV TGT+GEIRKHCA +N+
Sbjct: 325 TSMEKMGRINVKTGTEGEIRKHCAFLNS 352
>gb|AAA65637.1| peroxidase
Length = 328
Score = 106 bits (264), Expect = 2e-22
Identities = 54/88 (61%), Positives = 66/88 (74%), Gaps = 1/88 (1%)
Frame = -1
Query: 531 TLIEMDPGSRNTFDLGYYKQVLKRRGLFESDAALLNSSVTRSIITQQLRS-TQKFFLGFA 355
T++EMDPGS TFDL Y+K +LKRRGLF+SDAAL + T+S I Q + +FF FA
Sbjct: 241 TIVEMDPGSFKTFDLSYFKLLLKRRGLFQSDAALTTRTSTKSFIEQLVDGPLNEFFDEFA 300
Query: 354 KSMEKMGRINVLTGTKGEIRKHCALVNN 271
KSMEKMGR+ V TG+ GEIRKHCA VN+
Sbjct: 301 KSMEKMGRVEVKTGSAGEIRKHCAFVNS 328
>pir||T09218 peroxidase (EC 1.11.1.7) precursor prx10 - spinach (fragment)
Length = 322
Score = 105 bits (261), Expect = 4e-22
Identities = 54/87 (62%), Positives = 63/87 (72%), Gaps = 1/87 (1%)
Frame = -1
Query: 531 TLIEMDPGSRNTFDLGYYKQVLKRRGLFESDAALLNSSVTRSIITQQLRS-TQKFFLGFA 355
T++EMDPGS TFDL YYK +LKRRGLFESDAAL SS T S I + + + FF F+
Sbjct: 236 TIVEMDPGSHRTFDLSYYKLLLKRRGLFESDAALTKSSTTLSYIKELVNGPLETFFAEFS 295
Query: 354 KSMEKMGRINVLTGTKGEIRKHCALVN 274
KSM KMG + VLTG+ GEIRK CA VN
Sbjct: 296 KSMVKMGDVEVLTGSAGEIRKQCAFVN 322
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 453,968,649
Number of Sequences: 1393205
Number of extensions: 9532967
Number of successful extensions: 25051
Number of sequences better than 10.0: 419
Number of HSP's better than 10.0 without gapping: 24463
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24874
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17885181664
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)