Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003721A_C01 KMC003721A_c01
(816 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
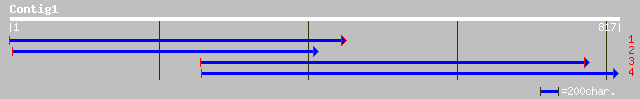
gb|AAC82382.1| putative odorant receptor LOR4 [Lampetra fluviati... 34 2.3
ref|XP_285081.1| similar to olfactory receptor MOR106-6 [Mus mus... 33 5.0
ref|XP_285082.1| similar to olfactory receptor MOR106-6 [Mus mus... 33 5.0
ref|NP_566493.1| expressed protein; protein id: At3g14770.1, sup... 33 6.6
ref|XP_139034.2| similar to olfactory receptor MOR106-6 [Mus mus... 32 8.6
>gb|AAC82382.1| putative odorant receptor LOR4 [Lampetra fluviatilis]
Length = 353
Score = 34.3 bits (77), Expect = 2.3
Identities = 24/78 (30%), Positives = 39/78 (49%), Gaps = 9/78 (11%)
Frame = -3
Query: 622 TLSTAGVLAGLPVKSSSFMGLQLRG*VNVLSLSLSPTRFSIG---------KY*IECCWT 470
TL A VL L + +S QL+ N+++LSL+ +G K +C +
Sbjct: 40 TLILATVLGNLLIITSIAYFRQLQTRTNIMALSLAVADLLVGLTVMPYSMMKAVYKCWFY 99
Query: 469 GLGFCNAWFFLSYVVTDA 416
G FCN +FL Y++T++
Sbjct: 100 GQFFCNLQYFLDYMLTNS 117
>ref|XP_285081.1| similar to olfactory receptor MOR106-6 [Mus musculus]
Length = 268
Score = 33.1 bits (74), Expect = 5.0
Identities = 30/116 (25%), Positives = 50/116 (42%), Gaps = 8/116 (6%)
Frame = -3
Query: 460 FCNAWFFLSYVVTDAW*LDFFATLENTRLLCLPSDHSI*NCLFWRISFVKCNRLES--VS 287
F +FF S+ T+ + F A + R L + C S + RL + V+
Sbjct: 59 FLQFYFFFSFGSTECF---FLAVMAFDRYLAI--------CRPLHYSSLMTGRLRNTLVT 107
Query: 286 FSYDCGFFF------LLSGLSIVTRRVRSFFSLDPG*FLFFACDR*PRVQLFYILI 137
+ GF + ++S +S R+ F DPG L AC R P +++F+ +I
Sbjct: 108 SCWVLGFLWFPVPIIIISQMSFCGSRIIDHFLCDPGPLLALACSRVPLIEVFWSII 163
>ref|XP_285082.1| similar to olfactory receptor MOR106-6 [Mus musculus]
Length = 254
Score = 33.1 bits (74), Expect = 5.0
Identities = 30/116 (25%), Positives = 50/116 (42%), Gaps = 8/116 (6%)
Frame = -3
Query: 460 FCNAWFFLSYVVTDAW*LDFFATLENTRLLCLPSDHSI*NCLFWRISFVKCNRLES--VS 287
F +FF S+ T+ + F A + R L + C S + RL + V+
Sbjct: 59 FLQFYFFFSFGSTECF---FLAVMAFDRYLAI--------CRPLHYSSLMTGRLRNTLVT 107
Query: 286 FSYDCGFFF------LLSGLSIVTRRVRSFFSLDPG*FLFFACDR*PRVQLFYILI 137
+ GF + ++S +S R+ F DPG L AC R P +++F+ +I
Sbjct: 108 SCWVLGFLWFPVPIIIISQMSFCGSRIIDHFLCDPGPLLALACSRVPLIEVFWSII 163
>ref|NP_566493.1| expressed protein; protein id: At3g14770.1, supported by cDNA:
gi_15809922, supported by cDNA: gi_17978878 [Arabidopsis
thaliana] gi|11994587|dbj|BAB02642.1| MtN3-like protein
[Arabidopsis thaliana] gi|15809923|gb|AAL06889.1|
AT3g14770/T21E2_2 [Arabidopsis thaliana]
gi|17978879|gb|AAL47411.1| AT3g14770/T21E2_2
[Arabidopsis thaliana]
Length = 236
Score = 32.7 bits (73), Expect = 6.6
Identities = 12/44 (27%), Positives = 22/44 (49%)
Frame = -3
Query: 148 YILIEISRGSWFGVPFLSHSTKIVSVIFKTHVRLYLCYFLIDVI 17
Y L+ W+G PF+SHS ++ + LCY ++ ++
Sbjct: 55 YALLNCLICLWYGTPFISHSNAMLMTVNSVGATFQLCYIILFIM 98
>ref|XP_139034.2| similar to olfactory receptor MOR106-6 [Mus musculus]
Length = 268
Score = 32.3 bits (72), Expect = 8.6
Identities = 28/109 (25%), Positives = 47/109 (42%), Gaps = 1/109 (0%)
Frame = -3
Query: 460 FCNAWFFLSYVVTDAW*LDFFATLENTRLLCLPSDH-SI*NCLFWRISFVKCNRLESVSF 284
F +FF S+ T+ + L A + +C P + S+ I + C L + F
Sbjct: 59 FLQFYFFFSFGSTECFFLAVMA-FDRYLAICRPLHYPSLMTRRLCNILVISCWVLGFLWF 117
Query: 283 SYDCGFFFLLSGLSIVTRRVRSFFSLDPG*FLFFACDR*PRVQLFYILI 137
++S +S R+ F DPG L AC R P +++F+ +I
Sbjct: 118 PVP---IIIISQMSFCGSRIIDHFLCDPGPLLALACSRAPLMEVFWTII 163
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 665,312,963
Number of Sequences: 1393205
Number of extensions: 14212168
Number of successful extensions: 25831
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 25118
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25817
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 42016716300
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)