Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003709A_C01 KMC003709A_c01
(586 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
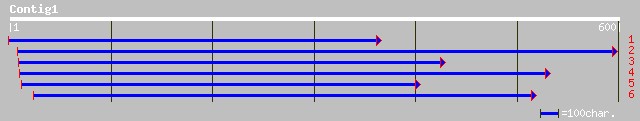
dbj|BAB89069.1| dynein light chain - like protein [Oryza sativa ... 132 3e-30
ref|NP_197511.1| dynein light chain - like protein; protein id: ... 130 9e-30
ref|NP_173736.1| dynein light subunit lc6, flagellar outer arm; ... 101 8e-21
gb|AAO51837.1| similar to Anthocidaris crassispina (Sea urchin).... 84 1e-15
sp|O02414|DYL1_ANTCR DYNEIN LIGHT CHAIN LC6, FLAGELLAR OUTER ARM... 83 2e-15
>dbj|BAB89069.1| dynein light chain - like protein [Oryza sativa (japonica
cultivar-group)] gi|20161709|dbj|BAB90626.1| dynein
light chain - like protein [Oryza sativa (japonica
cultivar-group)]
Length = 213
Score = 132 bits (332), Expect = 3e-30
Identities = 60/109 (55%), Positives = 87/109 (79%)
Frame = -1
Query: 565 RRSFCGSQVDLGDVFAINGAKMVSADMPPFMQIHAVDCARKAIDSMEKFTSKTLALSLKK 386
R+S ++++ +V ++ G K+++ADM PFMQ+HA CA+++ DS++KF+S+ LA +KK
Sbjct: 101 RKSMPEVEINMKEVVSVLGVKVMAADMSPFMQLHAFRCAKRSHDSLDKFSSRQLAHDVKK 160
Query: 385 EFDGVYGPAWHCIVGTSFGSFVTHSVGGFLYFSMDQKLYILLFKTTVQK 239
EFD VYGP WHCIVGTS+GSFVTH+ G FLYFSMD K+ ++LFKT ++K
Sbjct: 161 EFDKVYGPTWHCIVGTSYGSFVTHARGCFLYFSMD-KIIVMLFKTKIRK 208
>ref|NP_197511.1| dynein light chain - like protein; protein id: At5g20110.1
[Arabidopsis thaliana]
Length = 209
Score = 130 bits (328), Expect = 9e-30
Identities = 62/90 (68%), Positives = 75/90 (82%)
Frame = -1
Query: 505 KMVSADMPPFMQIHAVDCARKAIDSMEKFTSKTLALSLKKEFDGVYGPAWHCIVGTSFGS 326
++++ADMP FMQ HA CAR +DS+EKF+SK +A +LKKEFD YGPAWHCIVG+SFGS
Sbjct: 118 RILAADMPGFMQAHAFRCARMTLDSLEKFSSKHMAFNLKKEFDKGYGPAWHCIVGSSFGS 177
Query: 325 FVTHSVGGFLYFSMDQKLYILLFKTTVQKA 236
FVTHS G F+YFSMD KLY+LLFKT V+ A
Sbjct: 178 FVTHSTGCFIYFSMD-KLYVLLFKTKVRPA 206
>ref|NP_173736.1| dynein light subunit lc6, flagellar outer arm; protein id:
At1g23220.1, supported by cDNA: gi_17380801, supported
by cDNA: gi_20465366 [Arabidopsis thaliana]
gi|9295690|gb|AAF86996.1|AC005292_5 F26F24.7
[Arabidopsis thaliana] gi|17380802|gb|AAL36088.1|
putative dynein light subunit lc6, flagellar outer arm
[Arabidopsis thaliana] gi|20465367|gb|AAM20087.1|
putative dynein light subunit lc6, flagellar outer arm
[Arabidopsis thaliana]
Length = 129
Score = 101 bits (251), Expect = 8e-21
Identities = 54/105 (51%), Positives = 72/105 (68%), Gaps = 1/105 (0%)
Frame = -1
Query: 544 QVDLGDVFAINGAKMVSADMPPFMQIHAVDCARKAIDSME-KFTSKTLALSLKKEFDGVY 368
Q D D F + ++ ++DMP Q A +R+ +++ K +K LA +LKK+FD Y
Sbjct: 27 QQDQKDEFNV---RVRASDMPLPQQNRAFSLSREILNATPGKADNKRLAHALKKDFDSAY 83
Query: 367 GPAWHCIVGTSFGSFVTHSVGGFLYFSMDQKLYILLFKTTVQKAD 233
GPAWHCIVGTSFGS+VTHS GGFLYF +D K+Y+LLFKT V+ D
Sbjct: 84 GPAWHCIVGTSFGSYVTHSTGGFLYFQID-KVYVLLFKTAVEPLD 127
>gb|AAO51837.1| similar to Anthocidaris crassispina (Sea urchin). Dynein light
chain LC6, flagellar outer arm [Dictyostelium
discoideum]
Length = 91
Score = 84.0 bits (206), Expect = 1e-15
Identities = 42/81 (51%), Positives = 54/81 (65%)
Frame = -1
Query: 496 SADMPPFMQIHAVDCARKAIDSMEKFTSKTLALSLKKEFDGVYGPAWHCIVGTSFGSFVT 317
+ADMP FMQ A +C KA + E + +A+ +KKEFD Y P WHCIVG SFGSFVT
Sbjct: 12 NADMPDFMQQDATECTIKAFE--ETNIERDIAMIIKKEFDKKYSPTWHCIVGKSFGSFVT 69
Query: 316 HSVGGFLYFSMDQKLYILLFK 254
H F+YF+++ K +LLFK
Sbjct: 70 HETKNFIYFNIN-KHSVLLFK 89
>sp|O02414|DYL1_ANTCR DYNEIN LIGHT CHAIN LC6, FLAGELLAR OUTER ARM
gi|2208914|dbj|BAA20525.1| outer arm dynein LC6
[Anthocidaris crassispina]
Length = 89
Score = 83.2 bits (204), Expect = 2e-15
Identities = 45/87 (51%), Positives = 58/87 (65%), Gaps = 1/87 (1%)
Frame = -1
Query: 508 AKMVSADMPPFMQIHAVDCARKAIDSMEKFT-SKTLALSLKKEFDGVYGPAWHCIVGTSF 332
A + +ADMP MQ AVDCA +A+ EKF K +A +KKEFD Y P WHCIVG +F
Sbjct: 6 AVIKNADMPEDMQQDAVDCATQAL---EKFNIEKDIAAYIKKEFDKKYNPTWHCIVGRNF 62
Query: 331 GSFVTHSVGGFLYFSMDQKLYILLFKT 251
GS+VTH F+YF + Q + +LLFK+
Sbjct: 63 GSYVTHETKHFIYFYLGQ-VAVLLFKS 88
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 552,796,029
Number of Sequences: 1393205
Number of extensions: 12412343
Number of successful extensions: 31027
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 29889
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30929
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)