Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003692A_C01 KMC003692A_c01
(626 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
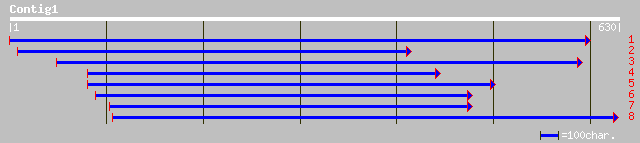
ref|NP_565461.1| putative N-acetyl-gamma-glutamyl-phosphate redu... 179 2e-44
gb|AAL24190.1| At2g19940/F6F22.3 [Arabidopsis thaliana] gi|19699... 179 2e-44
pir||A84583 hypothetical protein At2g19940 [imported] - Arabidop... 179 2e-44
gb|AAM92821.1| putative N-acetyl-gamma-glutamyl-phosphate reduct... 157 1e-37
gb|ZP_00053363.1| hypothetical protein [Magnetospirillum magneto... 130 1e-29
>ref|NP_565461.1| putative N-acetyl-gamma-glutamyl-phosphate reductase; protein id:
At2g19940.1, supported by cDNA: gi_16604367, supported
by cDNA: gi_19699207 [Arabidopsis thaliana]
gi|20197293|gb|AAC62122.2| putative
N-acetyl-gamma-glutamyl-phosphate reductase [Arabidopsis
thaliana]
Length = 359
Score = 179 bits (455), Expect = 2e-44
Identities = 85/111 (76%), Positives = 102/111 (91%)
Frame = -1
Query: 626 IPMSRGMQSTIYVEMAPGVRIEDLYEQLKLSYEDEKFVVLLEKGVIPRTHSVKGSNYCLI 447
+PM RGMQSTIYVEMAPGVR EDL++QLK SYEDE+FV +L++GV+PRTH+V+GSNYC +
Sbjct: 249 MPMIRGMQSTIYVEMAPGVRTEDLHQQLKTSYEDEEFVKVLDEGVVPRTHNVRGSNYCHM 308
Query: 446 NVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPEDLGLHYLPLFP 294
+VFPDRIPGRAIIISVIDNLVKGASGQALQNLN+++G+PE GL + PLFP
Sbjct: 309 SVFPDRIPGRAIIISVIDNLVKGASGQALQNLNIMLGYPETTGLLHQPLFP 359
>gb|AAL24190.1| At2g19940/F6F22.3 [Arabidopsis thaliana] gi|19699208|gb|AAL90970.1|
At2g19940/F6F22.3 [Arabidopsis thaliana]
Length = 401
Score = 179 bits (455), Expect = 2e-44
Identities = 85/111 (76%), Positives = 102/111 (91%)
Frame = -1
Query: 626 IPMSRGMQSTIYVEMAPGVRIEDLYEQLKLSYEDEKFVVLLEKGVIPRTHSVKGSNYCLI 447
+PM RGMQSTIYVEMAPGVR EDL++QLK SYEDE+FV +L++GV+PRTH+V+GSNYC +
Sbjct: 291 MPMIRGMQSTIYVEMAPGVRTEDLHQQLKTSYEDEEFVKVLDEGVVPRTHNVRGSNYCHM 350
Query: 446 NVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPEDLGLHYLPLFP 294
+VFPDRIPGRAIIISVIDNLVKGASGQALQNLN+++G+PE GL + PLFP
Sbjct: 351 SVFPDRIPGRAIIISVIDNLVKGASGQALQNLNIMLGYPETTGLLHQPLFP 401
>pir||A84583 hypothetical protein At2g19940 [imported] - Arabidopsis thaliana
Length = 389
Score = 179 bits (455), Expect = 2e-44
Identities = 85/111 (76%), Positives = 102/111 (91%)
Frame = -1
Query: 626 IPMSRGMQSTIYVEMAPGVRIEDLYEQLKLSYEDEKFVVLLEKGVIPRTHSVKGSNYCLI 447
+PM RGMQSTIYVEMAPGVR EDL++QLK SYEDE+FV +L++GV+PRTH+V+GSNYC +
Sbjct: 279 MPMIRGMQSTIYVEMAPGVRTEDLHQQLKTSYEDEEFVKVLDEGVVPRTHNVRGSNYCHM 338
Query: 446 NVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPEDLGLHYLPLFP 294
+VFPDRIPGRAIIISVIDNLVKGASGQALQNLN+++G+PE GL + PLFP
Sbjct: 339 SVFPDRIPGRAIIISVIDNLVKGASGQALQNLNIMLGYPETTGLLHQPLFP 389
>gb|AAM92821.1| putative N-acetyl-gamma-glutamyl-phosphate reductase [Oryza sativa
(japonica cultivar-group)]
Length = 416
Score = 157 bits (396), Expect = 1e-37
Identities = 78/111 (70%), Positives = 88/111 (79%)
Frame = -1
Query: 626 IPMSRGMQSTIYVEMAPGVRIEDLYEQLKLSYEDEKFVVLLEKGVIPRTHSVKGSNYCLI 447
I M RGMQST++VEMAPGV DLY+ LK +YE E+FV LL +P T V GSNYC +
Sbjct: 306 ICMKRGMQSTMFVEMAPGVTAGDLYQHLKSTYEGEEFVKLLHGSTVPHTRHVVGSNYCFM 365
Query: 446 NVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPEDLGLHYLPLFP 294
NVF DRIPGRAIIISVIDNLVKGASGQA+QNLNL+MG PE+ GL Y PLFP
Sbjct: 366 NVFEDRIPGRAIIISVIDNLVKGASGQAVQNLNLMMGLPENRGLQYQPLFP 416
>gb|ZP_00053363.1| hypothetical protein [Magnetospirillum magnetotacticum]
Length = 275
Score = 130 bits (328), Expect = 1e-29
Identities = 65/111 (58%), Positives = 79/111 (70%)
Frame = -1
Query: 626 IPMSRGMQSTIYVEMAPGVRIEDLYEQLKLSYEDEKFVVLLEKGVIPRTHSVKGSNYCLI 447
IPMSRGM STIYV+ G + DL L ++ E FV ++ +GV+P T V+GSN+CLI
Sbjct: 165 IPMSRGMLSTIYVKTKNGAKAADLRAHLSKTFGGEPFVRVVPEGVVPHTRHVRGSNHCLI 224
Query: 446 NVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPEDLGLHYLPLFP 294
N F DR+PGR II SVIDNLVKGASGQALQN+NL+ G PE L P+FP
Sbjct: 225 NAFDDRVPGRIIITSVIDNLVKGASGQALQNMNLMFGLPEATALAQQPMFP 275
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 509,302,328
Number of Sequences: 1393205
Number of extensions: 10430991
Number of successful extensions: 20835
Number of sequences better than 10.0: 159
Number of HSP's better than 10.0 without gapping: 20274
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20784
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25586195130
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)