Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003685A_C01 KMC003685A_c01
(720 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
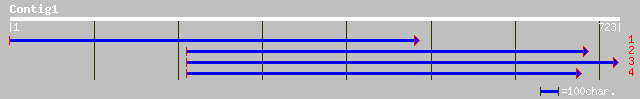
dbj|BAC42111.1| unknown protein [Arabidopsis thaliana] 73 3e-12
ref|NP_176591.1| hypothetical protein; protein id: At1g64080.1 [... 73 3e-12
dbj|BAB32722.1| P0492F05.24 [Oryza sativa (japonica cultivar-gro... 69 7e-11
ref|NP_200099.1| putative protein; protein id: At5g52870.1, supp... 54 2e-06
gb|EAA00112.1| agCP9331 [Anopheles gambiae str. PEST] 37 0.28
>dbj|BAC42111.1| unknown protein [Arabidopsis thaliana]
Length = 411
Score = 73.2 bits (178), Expect = 3e-12
Identities = 38/65 (58%), Positives = 48/65 (73%), Gaps = 5/65 (7%)
Frame = -3
Query: 715 ASSAVAASPPAAAA-----ESSKRRDDSLLQQQDAIQGAILHCQRSFNASRGCESSQLPR 551
+SS+ A +P + A S+RRDDSLLQQQD+IQ AILHC+RSFN+SR + S LPR
Sbjct: 342 SSSSTATTPSSTTATVTTQSESRRRDDSLLQQQDSIQSAILHCKRSFNSSRDKDPSVLPR 401
Query: 550 SVSDP 536
SVS+P
Sbjct: 402 SVSEP 406
>ref|NP_176591.1| hypothetical protein; protein id: At1g64080.1 [Arabidopsis
thaliana] gi|25404922|pir||E96665 protein F22C12.16
[imported] - Arabidopsis thaliana
gi|6692100|gb|AAF24565.1|AC007764_7 F22C12.16
[Arabidopsis thaliana]
Length = 411
Score = 73.2 bits (178), Expect = 3e-12
Identities = 38/65 (58%), Positives = 48/65 (73%), Gaps = 5/65 (7%)
Frame = -3
Query: 715 ASSAVAASPPAAAA-----ESSKRRDDSLLQQQDAIQGAILHCQRSFNASRGCESSQLPR 551
+SS+ A +P + A S+RRDDSLLQQQD+IQ AILHC+RSFN+SR + S LPR
Sbjct: 342 SSSSTATTPSSTTATVTTQSESRRRDDSLLQQQDSIQSAILHCKRSFNSSRDKDPSVLPR 401
Query: 550 SVSDP 536
SVS+P
Sbjct: 402 SVSEP 406
>dbj|BAB32722.1| P0492F05.24 [Oryza sativa (japonica cultivar-group)]
gi|20804415|dbj|BAB92113.1| P0443E07.16 [Oryza sativa
(japonica cultivar-group)]
Length = 411
Score = 68.9 bits (167), Expect = 7e-11
Identities = 39/67 (58%), Positives = 46/67 (68%), Gaps = 7/67 (10%)
Frame = -3
Query: 718 SASSAVAASP-------PAAAAESSKRRDDSLLQQQDAIQGAILHCQRSFNASRGCESSQ 560
SASSAVAA+P P+ A + +RRDDSLLQ QD IQ AI HC+RSFNAS+ S
Sbjct: 326 SASSAVAAAPSPSLPPPPSTAGQQPQRRDDSLLQLQDGIQSAIAHCKRSFNASKVGSESP 385
Query: 559 LPRSVSD 539
L RS+SD
Sbjct: 386 LLRSMSD 392
>ref|NP_200099.1| putative protein; protein id: At5g52870.1, supported by cDNA:
gi_15028096, supported by cDNA: gi_20258914 [Arabidopsis
thaliana] gi|10177101|dbj|BAB10435.1|
gb|AAF24565.1~gene_id:MXC20.9~similar to unknown protein
[Arabidopsis thaliana] gi|15028097|gb|AAK76579.1|
unknown protein [Arabidopsis thaliana]
gi|20258915|gb|AAM14151.1| unknown protein [Arabidopsis
thaliana]
Length = 326
Score = 53.9 bits (128), Expect = 2e-06
Identities = 37/82 (45%), Positives = 47/82 (57%)
Frame = -3
Query: 718 SASSAVAASPPAAAAESSKRRDDSLLQQQDAIQGAILHCQRSFNASRGCESSQLPRSVSD 539
SAS+A+ PA R D+SL QQD IQ AILHC++SF+ SR ESS L RS S+
Sbjct: 229 SASAAIGGMSPA------NRIDESLQVQQDGIQSAILHCKKSFHGSR--ESSLLSRSTSE 280
Query: 538 PLHDVSSELSRKSTTEGKDKRL 473
SS + ST+ +D L
Sbjct: 281 -----SSSQEKLSTSSSEDSYL 297
>gb|EAA00112.1| agCP9331 [Anopheles gambiae str. PEST]
Length = 1922
Score = 37.0 bits (84), Expect = 0.28
Identities = 26/81 (32%), Positives = 38/81 (46%)
Frame = -3
Query: 694 SPPAAAAESSKRRDDSLLQQQDAIQGAILHCQRSFNASRGCESSQLPRSVSDPLHDVSSE 515
+P AA SSKR DD +D+ + C+ N S SS S SD +E
Sbjct: 1418 TPEAAVKSSSKRNDDQSSDSEDS-SDSDSSCECGSNCSCSSSSSNSSSSSSDDSDSSDNE 1476
Query: 514 LSRKSTTEGKDKRLPLKNEEK 452
+RK + +G D+ P K E++
Sbjct: 1477 DARKGSKKGTDQ--PKKEEDE 1495
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 586,931,235
Number of Sequences: 1393205
Number of extensions: 12303818
Number of successful extensions: 63820
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 46869
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 61366
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33499052993
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)