Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003678A_C01 KMC003678A_c01
(763 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
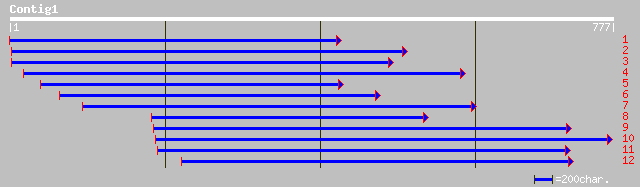
dbj|BAC42351.1| unknown protein [Arabidopsis thaliana] 191 1e-47
ref|NP_195382.1| putative protein; protein id: At4g36640.1 [Arab... 191 1e-47
ref|NP_177653.1| unknown protein; protein id: At1g75170.1, suppo... 187 1e-46
ref|NP_196098.1| putative protein; protein id: At5g04780.1 [Arab... 171 8e-42
pir||D86354 F16L1.9 protein - Arabidopsis thaliana gi|9454532|gb... 112 4e-24
>dbj|BAC42351.1| unknown protein [Arabidopsis thaliana]
Length = 294
Score = 191 bits (484), Expect = 1e-47
Identities = 90/141 (63%), Positives = 112/141 (78%)
Frame = -2
Query: 762 IDFTGFSLGTDLSPTTARDIIHILQNHYPERLAIAFLFNPPRIFQAFYKAVKYFLDPKTA 583
IDFTG+S+ + T R+IIHILQN+YPERL IAFL+NPPR+FQA Y+A KYFLDP+TA
Sbjct: 146 IDFTGWSMAVNPPMKTTREIIHILQNYYPERLGIAFLYNPPRLFQAVYRAAKYFLDPRTA 205
Query: 582 QKVKFVYPNNKDSVELMKSLFDIDNLPSEFGGKATLKYDHEEFSRLMTEDDVKTAKYWGF 403
+KVKFVYP +K S ELM + FD++NLP EFGG+ATL+YDHE+FSR M EDD+KTAKYWG
Sbjct: 206 EKVKFVYPKDKASDELMATHFDVENLPKEFGGEATLEYDHEDFSRQMYEDDLKTAKYWGL 265
Query: 402 DDDKPFTTMNGHIGAEVAPEP 340
+ K + NG ++V PEP
Sbjct: 266 -EGKHYPKTNGFSPSDVVPEP 285
>ref|NP_195382.1| putative protein; protein id: At4g36640.1 [Arabidopsis thaliana]
gi|25407749|pir||F85432 hypothetical protein AT4g36640
[imported] - Arabidopsis thaliana
gi|4006899|emb|CAB16829.1| putative protein [Arabidopsis
thaliana] gi|7270612|emb|CAB80330.1| putative protein
[Arabidopsis thaliana]
Length = 294
Score = 191 bits (484), Expect = 1e-47
Identities = 90/141 (63%), Positives = 112/141 (78%)
Frame = -2
Query: 762 IDFTGFSLGTDLSPTTARDIIHILQNHYPERLAIAFLFNPPRIFQAFYKAVKYFLDPKTA 583
IDFTG+S+ + T R+IIHILQN+YPERL IAFL+NPPR+FQA Y+A KYFLDP+TA
Sbjct: 146 IDFTGWSMAVNPPMKTTREIIHILQNYYPERLGIAFLYNPPRLFQAVYRAAKYFLDPRTA 205
Query: 582 QKVKFVYPNNKDSVELMKSLFDIDNLPSEFGGKATLKYDHEEFSRLMTEDDVKTAKYWGF 403
+KVKFVYP +K S ELM + FD++NLP EFGG+ATL+YDHE+FSR M EDD+KTAKYWG
Sbjct: 206 EKVKFVYPKDKASDELMTTHFDVENLPKEFGGEATLEYDHEDFSRQMYEDDLKTAKYWGL 265
Query: 402 DDDKPFTTMNGHIGAEVAPEP 340
+ K + NG ++V PEP
Sbjct: 266 -EGKHYPKTNGFSPSDVVPEP 285
>ref|NP_177653.1| unknown protein; protein id: At1g75170.1, supported by cDNA:
gi_17979168, supported by cDNA: gi_20259124 [Arabidopsis
thaliana] gi|25406417|pir||H96781 unknown protein
F22H5.20 [imported] - Arabidopsis thaliana
gi|10092270|gb|AAG12683.1|AC025814_7 unknown protein;
51719-50438 [Arabidopsis thaliana]
gi|17979169|gb|AAL49780.1| unknown protein [Arabidopsis
thaliana] gi|20259125|gb|AAM14278.1| unknown protein
[Arabidopsis thaliana]
Length = 296
Score = 187 bits (476), Expect = 1e-46
Identities = 88/148 (59%), Positives = 114/148 (76%), Gaps = 2/148 (1%)
Frame = -2
Query: 762 IDFTGFSLGTDLSPTTARDIIHILQNHYPERLAIAFLFNPPRIFQAFYKAVKYFLDPKTA 583
IDFTG+S+ T + +AR+ I+ILQNHYPERLA+AFL+NPPR+F+AF+K VKYF+D KT
Sbjct: 149 IDFTGWSMSTSVPIKSARETINILQNHYPERLAVAFLYNPPRLFEAFWKIVKYFIDAKTF 208
Query: 582 QKVKFVYPNNKDSVELMKSLFDIDNLPSEFGGKATLKYDHEEFSRLMTEDDVKTAKYWGF 403
KVKFVYP N +SVELM + FD +NLP+EFGGKA L+Y++EEFS+ M +DDVKTA +WG
Sbjct: 209 VKVKFVYPKNSESVELMSTFFDEENLPTEFGGKALLQYNYEEFSKQMNQDDVKTANFWGL 268
Query: 402 --DDDKPFTTMNGHIGAEVAPEPVPVTP 325
++ NG GAE+APEP+ P
Sbjct: 269 GHSNNNQLHASNGFSGAEIAPEPIQNHP 296
>ref|NP_196098.1| putative protein; protein id: At5g04780.1 [Arabidopsis thaliana]
gi|11358100|pir||T48474 hypothetical protein T1E3.140 -
Arabidopsis thaliana gi|7413540|emb|CAB86020.1| putative
protein [Arabidopsis thaliana] gi|9758453|dbj|BAB08982.1|
selenium-binding protein-like [Arabidopsis thaliana]
Length = 864
Score = 171 bits (434), Expect = 8e-42
Identities = 84/148 (56%), Positives = 109/148 (72%), Gaps = 2/148 (1%)
Frame = -2
Query: 762 IDFTGFSLGTDLSPTTARDIIHILQNHYPERLAIAFLFNPPRIFQAFYKAVKYFLDPKTA 583
IDFTG+S+ T + +AR+ I+ILQNHYPERLA+AFL+NPPR+F+AF+K KT
Sbjct: 722 IDFTGWSMSTSVPIKSARETINILQNHYPERLAVAFLYNPPRLFEAFWKE-----HAKTF 776
Query: 582 QKVKFVYPNNKDSVELMKSLFDIDNLPSEFGGKATLKYDHEEFSRLMTEDDVKTAKYWGF 403
KVKFVYP N++SVELM + FD +NLP+EFGGKA L+Y++EEFS+ M +DDVKTA +WG
Sbjct: 777 VKVKFVYPKNQESVELMSTFFDEENLPTEFGGKALLQYNYEEFSKQMNQDDVKTANFWGL 836
Query: 402 --DDDKPFTTMNGHIGAEVAPEPVPVTP 325
++ NG GAE+APEPV P
Sbjct: 837 CNSNNNQLHASNGFSGAEIAPEPVQTHP 864
>pir||D86354 F16L1.9 protein - Arabidopsis thaliana
gi|9454532|gb|AAF87855.1|AC073942_9 Contains similarity
to a KIAA0420 protein from Homo sapiens gi|2887415 and
contains a CRAL/TRIO PF|00650 domain. [Arabidopsis
thaliana]
Length = 314
Score = 112 bits (281), Expect = 4e-24
Identities = 54/114 (47%), Positives = 83/114 (72%), Gaps = 1/114 (0%)
Frame = -2
Query: 762 IDFTGFSLGTDLSPTTARDIIHILQNHYPERLAIAFLFNPPRIFQAFYKAVKYFLDPKTA 583
IDF GF++ + +S +R+ H+LQ HYPERL +A ++NPP+IF++FYK VK FL+PKT+
Sbjct: 151 IDFHGFNM-SHISLKVSRETAHVLQEHYPERLGLAIVYNPPKIFESFYKMVKPFLEPKTS 209
Query: 582 QKVKFVYPNNKDSVELMKSLFDIDNLPSEFGGK-ATLKYDHEEFSRLMTEDDVK 424
KVKFVY ++ S +L++ LFD++ L FGGK + ++ E+++ M EDD+K
Sbjct: 210 NKVKFVYSDDNLSNKLLEDLFDMEQLEVAFGGKNSDAGFNFEKYAERMREDDLK 263
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 620,817,422
Number of Sequences: 1393205
Number of extensions: 13197935
Number of successful extensions: 34001
Number of sequences better than 10.0: 158
Number of HSP's better than 10.0 without gapping: 32403
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33956
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 36974710344
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)