Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003671A_C01 KMC003671A_c01
(652 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
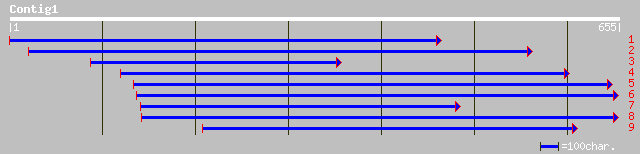
ref|NP_567405.1| putative protein; protein id: At4g13430.1, supp... 223 1e-57
pir||T06300 hypothetical protein T9E8.170 - Arabidopsis thaliana... 223 1e-57
ref|ZP_00074911.1| hypothetical protein [Trichodesmium erythraeu... 171 1e-41
ref|NP_661514.1| 3-isopropylmalate dehydratase, large subunit, p... 115 4e-25
ref|NP_614723.1| 3-isopropylmalate dehydratase large subunit [Me... 88 9e-17
>ref|NP_567405.1| putative protein; protein id: At4g13430.1, supported by cDNA:
gi_15027970 [Arabidopsis thaliana]
gi|15027971|gb|AAK76516.1| unknown protein [Arabidopsis
thaliana] gi|21436051|gb|AAM51226.1| unknown protein
[Arabidopsis thaliana]
Length = 509
Score = 223 bits (569), Expect = 1e-57
Identities = 103/112 (91%), Positives = 109/112 (96%)
Frame = -3
Query: 650 VKVPTFLVPATQKVWMDVYSLPVPGSGGKTCSQIFEEAGCDTPASPSCGACLGGPKDTYA 471
VKVPTFLVPATQKVWMDVY+LPVPG+GGKTC+QIFEEAGCDTPASPSCGACLGGP DTYA
Sbjct: 398 VKVPTFLVPATQKVWMDVYALPVPGAGGKTCAQIFEEAGCDTPASPSCGACLGGPADTYA 457
Query: 470 RMNEPKVCVSTTNRNFPGRMGHKEGEIYLASPYTAAASALTGYVTDPREFLQ 315
R+NEP+VCVSTTNRNFPGRMGHKEG+IYLASPYTAAASALTG V DPREFLQ
Sbjct: 458 RLNEPQVCVSTTNRNFPGRMGHKEGQIYLASPYTAAASALTGRVADPREFLQ 509
>pir||T06300 hypothetical protein T9E8.170 - Arabidopsis thaliana
gi|4584548|emb|CAB40778.1| putative protein [Arabidopsis
thaliana] gi|7268046|emb|CAB78385.1| putative protein
[Arabidopsis thaliana]
Length = 509
Score = 223 bits (569), Expect = 1e-57
Identities = 103/112 (91%), Positives = 109/112 (96%)
Frame = -3
Query: 650 VKVPTFLVPATQKVWMDVYSLPVPGSGGKTCSQIFEEAGCDTPASPSCGACLGGPKDTYA 471
VKVPTFLVPATQKVWMDVY+LPVPG+GGKTC+QIFEEAGCDTPASPSCGACLGGP DTYA
Sbjct: 398 VKVPTFLVPATQKVWMDVYALPVPGAGGKTCAQIFEEAGCDTPASPSCGACLGGPADTYA 457
Query: 470 RMNEPKVCVSTTNRNFPGRMGHKEGEIYLASPYTAAASALTGYVTDPREFLQ 315
R+NEP+VCVSTTNRNFPGRMGHKEG+IYLASPYTAAASALTG V DPREFLQ
Sbjct: 458 RLNEPQVCVSTTNRNFPGRMGHKEGQIYLASPYTAAASALTGRVADPREFLQ 509
>ref|ZP_00074911.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 444
Score = 171 bits (432), Expect = 1e-41
Identities = 79/111 (71%), Positives = 96/111 (86%)
Frame = -3
Query: 650 VKVPTFLVPATQKVWMDVYSLPVPGSGGKTCSQIFEEAGCDTPASPSCGACLGGPKDTYA 471
VKVPT+LVPATQKV+ D++ + GKT S+IF +AGC PA+PSC ACLGGPKDT+
Sbjct: 337 VKVPTYLVPATQKVYEDLFIIK---HDGKTLSEIFLDAGCIEPAAPSCAACLGGPKDTFG 393
Query: 470 RMNEPKVCVSTTNRNFPGRMGHKEGEIYLASPYTAAASALTGYVTDPREFL 318
R+NEP++CVSTTNRNFPGRMG+K+ ++YLASPYTAAASALTGYVTDPREF+
Sbjct: 394 RLNEPEICVSTTNRNFPGRMGNKQAQVYLASPYTAAASALTGYVTDPREFM 444
>ref|NP_661514.1| 3-isopropylmalate dehydratase, large subunit, putative [Chlorobium
tepidum TLS] gi|21646552|gb|AAM71856.1|
3-isopropylmalate dehydratase, large subunit, putative
[Chlorobium tepidum TLS]
Length = 431
Score = 115 bits (289), Expect = 4e-25
Identities = 65/111 (58%), Positives = 73/111 (65%)
Frame = -3
Query: 650 VKVPTFLVPATQKVWMDVYSLPVPGSGGKTCSQIFEEAGCDTPASPSCGACLGGPKDTYA 471
V V T +VPAT V L G+T IFEEAGC+ A PSC ACLGGP DT
Sbjct: 325 VAVTTNIVPATVLV---ASQLETEMYDGQTLRHIFEEAGCNI-ALPSCAACLGGPSDTVG 380
Query: 470 RMNEPKVCVSTTNRNFPGRMGHKEGEIYLASPYTAAASALTGYVTDPREFL 318
R + V VSTTNRNFPGRMG K +YLASP TAAASA+TG +TDPR+FL
Sbjct: 381 RSVDNDVVVSTTNRNFPGRMGSKFASVYLASPLTAAASAITGKLTDPRDFL 431
>ref|NP_614723.1| 3-isopropylmalate dehydratase large subunit [Methanopyrus kandleri
AV19] gi|22001713|sp|Q8TVF2|LE21_METKA 3-isopropylmalate
dehydratase large subunit 1 (Isopropylmalate isomerase
1) (Alpha-IPM isomerase 1) (IPMI 1)
gi|19888106|gb|AAM02653.1| 3-isopropylmalate dehydratase
large subunit [Methanopyrus kandleri AV19]
Length = 418
Score = 88.2 bits (217), Expect = 9e-17
Identities = 44/79 (55%), Positives = 55/79 (68%)
Frame = -3
Query: 554 QIFEEAGCDTPASPSCGACLGGPKDTYARMNEPKVCVSTTNRNFPGRMGHKEGEIYLASP 375
++ EAG P+CG CLGG A E + CV+T+NRNFPGRMGH+E E+YLASP
Sbjct: 344 EVLHEAGA-LICPPNCGPCLGGHMGVLA---EGERCVATSNRNFPGRMGHRESEVYLASP 399
Query: 374 YTAAASALTGYVTDPREFL 318
TAAASA+ G +TDPR +L
Sbjct: 400 ATAAASAIEGEITDPRPYL 418
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 565,006,045
Number of Sequences: 1393205
Number of extensions: 12623867
Number of successful extensions: 34941
Number of sequences better than 10.0: 321
Number of HSP's better than 10.0 without gapping: 32679
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34844
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27860523586
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)