Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003665A_C01 KMC003665A_c01
(684 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
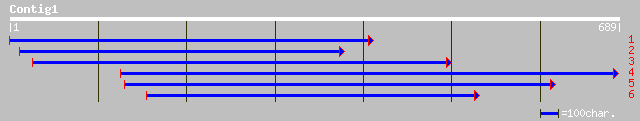
ref|NP_195828.1| putative protein; protein id: At5g02080.1 [Arab... 93 2e-22
ref|NP_563904.1| hypothetical protein; protein id: At1g12350.1 [... 92 7e-18
dbj|BAC42714.1| unknown protein [Arabidopsis thaliana] 92 7e-18
gb|AAF79631.1|AC025416_5 F5O11.7 [Arabidopsis thaliana] 92 7e-18
ref|NP_732401.1| CG5629-PA [Drosophila melanogaster] gi|24648134... 62 1e-10
>ref|NP_195828.1| putative protein; protein id: At5g02080.1 [Arabidopsis thaliana]
gi|11358399|pir||T48229 hypothetical protein T7H20.130 -
Arabidopsis thaliana gi|7340682|emb|CAB82981.1| putative
protein [Arabidopsis thaliana]
Length = 270
Score = 93.2 bits (230), Expect(2) = 2e-22
Identities = 46/59 (77%), Positives = 50/59 (83%)
Frame = -2
Query: 677 LVQVPKMLSVPRKDWAPLAFCVSFKLETDSSILLTKAGTALEKYKMHAVVANELSTRKE 501
L QVPKMLSV R +WAP AFC+SFKLETDS IL+ KA AL KYK+HAVVANELSTRKE
Sbjct: 167 LAQVPKMLSVLRSNWAPKAFCISFKLETDSKILMEKATKALRKYKVHAVVANELSTRKE 225
Score = 34.7 bits (78), Expect(2) = 2e-22
Identities = 17/45 (37%), Positives = 28/45 (61%)
Frame = -3
Query: 505 RSQVVVVTGAEKIKVQRDKSMSVNDVEDPLINLLSERHGPYIEAS 371
+ +VVVV+ + + V+R+ + VED LI L+ +RH YI+ S
Sbjct: 224 KEEVVVVSSSGNVVVRRECDKPESFVEDNLIRLIVDRHSTYIKES 268
>ref|NP_563904.1| hypothetical protein; protein id: At1g12350.1 [Arabidopsis
thaliana]
Length = 319
Score = 92.0 bits (227), Expect = 7e-18
Identities = 45/59 (76%), Positives = 50/59 (84%)
Frame = -2
Query: 677 LVQVPKMLSVPRKDWAPLAFCVSFKLETDSSILLTKAGTALEKYKMHAVVANELSTRKE 501
L QVPKMLS+ R +WAP AFC+SFKLETDS ILL KA AL+KYK+HAVVANEL TRKE
Sbjct: 216 LAQVPKMLSILRSNWAPKAFCISFKLETDSKILLEKATKALQKYKVHAVVANELLTRKE 274
Score = 40.8 bits (94), Expect = 0.018
Identities = 24/71 (33%), Positives = 42/71 (58%), Gaps = 6/71 (8%)
Frame = -3
Query: 565 VQLLKNTRCMQLLQMNSQLA------RSQVVVVTGAEKIKVQRDKSMSVNDVEDPLINLL 404
+ L K T+ +Q ++++ +A + +VVVV+ + + V+RD + + VED LI LL
Sbjct: 247 ILLEKATKALQKYKVHAVVANELLTRKEEVVVVSSSGNVVVRRDSNKPESIVEDNLIRLL 306
Query: 403 SERHGPYIEAS 371
+RH YI+ S
Sbjct: 307 VDRHSTYIKES 317
>dbj|BAC42714.1| unknown protein [Arabidopsis thaliana]
Length = 317
Score = 92.0 bits (227), Expect = 7e-18
Identities = 45/59 (76%), Positives = 50/59 (84%)
Frame = -2
Query: 677 LVQVPKMLSVPRKDWAPLAFCVSFKLETDSSILLTKAGTALEKYKMHAVVANELSTRKE 501
L QVPKMLS+ R +WAP AFC+SFKLETDS ILL KA AL+KYK+HAVVANEL TRKE
Sbjct: 214 LAQVPKMLSILRSNWAPKAFCISFKLETDSKILLEKATKALQKYKVHAVVANELLTRKE 272
Score = 40.8 bits (94), Expect = 0.018
Identities = 24/71 (33%), Positives = 42/71 (58%), Gaps = 6/71 (8%)
Frame = -3
Query: 565 VQLLKNTRCMQLLQMNSQLA------RSQVVVVTGAEKIKVQRDKSMSVNDVEDPLINLL 404
+ L K T+ +Q ++++ +A + +VVVV+ + + V+RD + + VED LI LL
Sbjct: 245 ILLEKATKALQKYKVHAVVANELLTRKEEVVVVSSSGNVVVRRDSNKPESIVEDNLIRLL 304
Query: 403 SERHGPYIEAS 371
+RH YI+ S
Sbjct: 305 VDRHSTYIKES 315
>gb|AAF79631.1|AC025416_5 F5O11.7 [Arabidopsis thaliana]
Length = 455
Score = 92.0 bits (227), Expect = 7e-18
Identities = 45/59 (76%), Positives = 50/59 (84%)
Frame = -2
Query: 677 LVQVPKMLSVPRKDWAPLAFCVSFKLETDSSILLTKAGTALEKYKMHAVVANELSTRKE 501
L QVPKMLS+ R +WAP AFC+SFKLETDS ILL KA AL+KYK+HAVVANEL TRKE
Sbjct: 352 LAQVPKMLSILRSNWAPKAFCISFKLETDSKILLEKATKALQKYKVHAVVANELLTRKE 410
Score = 40.8 bits (94), Expect = 0.018
Identities = 24/71 (33%), Positives = 42/71 (58%), Gaps = 6/71 (8%)
Frame = -3
Query: 565 VQLLKNTRCMQLLQMNSQLA------RSQVVVVTGAEKIKVQRDKSMSVNDVEDPLINLL 404
+ L K T+ +Q ++++ +A + +VVVV+ + + V+RD + + VED LI LL
Sbjct: 383 ILLEKATKALQKYKVHAVVANELLTRKEEVVVVSSSGNVVVRRDSNKPESIVEDNLIRLL 442
Query: 403 SERHGPYIEAS 371
+RH YI+ S
Sbjct: 443 VDRHSTYIKES 453
>ref|NP_732401.1| CG5629-PA [Drosophila melanogaster] gi|24648134|ref|NP_650784.2|
CG5629-PB [Drosophila melanogaster]
gi|4972686|gb|AAD34738.1| unknown [Drosophila
melanogaster] gi|7300482|gb|AAF55637.1| CG5629-PB
[Drosophila melanogaster] gi|23171702|gb|AAN13797.1|
CG5629-PA [Drosophila melanogaster]
Length = 313
Score = 62.0 bits (149), Expect(2) = 1e-10
Identities = 32/55 (58%), Positives = 37/55 (67%)
Frame = -2
Query: 668 VPKMLSVPRKDWAPLAFCVSFKLETDSSILLTKAGTALEKYKMHAVVANELSTRK 504
VPKML+ W P AF VSFKLETD S+L+ KA +L KYK V+AN L TRK
Sbjct: 209 VPKMLAPLASLWVPHAFVVSFKLETDESLLIVKARDSLNKYKHKLVIANVLQTRK 263
Score = 26.2 bits (56), Expect(2) = 1e-10
Identities = 11/47 (23%), Positives = 30/47 (63%), Gaps = 2/47 (4%)
Frame = -3
Query: 514 QLARSQVVVVTGAE--KIKVQRDKSMSVNDVEDPLINLLSERHGPYI 380
Q + +VV VT + ++ + R++++ ++E+P++ + ++HG +I
Sbjct: 260 QTRKHRVVFVTPTDSYELHLTREQTLQGLEIEEPIVADVVQKHGEFI 306
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 539,467,998
Number of Sequences: 1393205
Number of extensions: 11209626
Number of successful extensions: 24722
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 23924
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24720
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30552968016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)