Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003658A_C01 KMC003658A_c01
(555 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
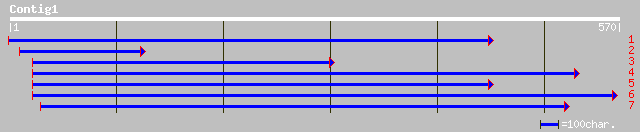
dbj|BAA95830.1| EST AU082557(R0845) corresponds to a region of t... 171 7e-42
dbj|BAA95820.1| EST AU082557(R0845) corresponds to a region of t... 171 7e-42
gb|AAF74980.1|AF082890_1 cystathionine beta-lyase [Solanum tuber... 160 7e-39
gb|AAL36374.1| putative cystathionine beta-lyase precursor CBL [... 157 6e-38
ref|NP_191264.1| CYSTATHIONINE BETA-LYASE PRECURSOR (CBL); prote... 157 6e-38
>dbj|BAA95830.1| EST AU082557(R0845) corresponds to a region of the predicted
gene.~Similar to Arabidopsis thaliana cystathionine
beta-lyase precursor (P53780) [Oryza sativa (japonica
cultivar-group)]
Length = 383
Score = 171 bits (432), Expect = 7e-42
Identities = 84/96 (87%), Positives = 93/96 (96%)
Frame = -2
Query: 551 LHYSQAKGTGSVLSFLTGSLALSKHIVETTKYYSITVSFGSVKSLISLPCFMSHASIPAA 372
LHYSQAKG GSVLSFLTGSLALSKH+VETTKY+++TVSFGSVKSLISLPCFMSHASIP+A
Sbjct: 287 LHYSQAKGAGSVLSFLTGSLALSKHVVETTKYFNVTVSFGSVKSLISLPCFMSHASIPSA 346
Query: 371 VREVRGLTEDLVRISVGIEDANDLIADLNHAFRTGP 264
VRE RGLT+DLVRISVGIEDA+DLIADL+HA R+GP
Sbjct: 347 VREERGLTDDLVRISVGIEDADDLIADLDHALRSGP 382
>dbj|BAA95820.1| EST AU082557(R0845) corresponds to a region of the predicted
gene.~Similar to Arabidopsis thaliana cystathionine
beta-lyase precursor (P53780) [Oryza sativa (japonica
cultivar-group)]
Length = 475
Score = 171 bits (432), Expect = 7e-42
Identities = 84/96 (87%), Positives = 93/96 (96%)
Frame = -2
Query: 551 LHYSQAKGTGSVLSFLTGSLALSKHIVETTKYYSITVSFGSVKSLISLPCFMSHASIPAA 372
LHYSQAKG GSVLSFLTGSLALSKH+VETTKY+++TVSFGSVKSLISLPCFMSHASIP+A
Sbjct: 379 LHYSQAKGAGSVLSFLTGSLALSKHVVETTKYFNVTVSFGSVKSLISLPCFMSHASIPSA 438
Query: 371 VREVRGLTEDLVRISVGIEDANDLIADLNHAFRTGP 264
VRE RGLT+DLVRISVGIEDA+DLIADL+HA R+GP
Sbjct: 439 VREERGLTDDLVRISVGIEDADDLIADLDHALRSGP 474
>gb|AAF74980.1|AF082890_1 cystathionine beta-lyase [Solanum tuberosum]
Length = 471
Score = 160 bits (406), Expect = 7e-39
Identities = 80/96 (83%), Positives = 86/96 (89%)
Frame = -2
Query: 551 LHYSQAKGTGSVLSFLTGSLALSKHIVETTKYYSITVSFGSVKSLISLPCFMSHASIPAA 372
LH+SQA G GSVLSFLTGSLALSKH+ E TKY+SITVSFGSVKSLISLPCFMSHASIP
Sbjct: 375 LHFSQATGAGSVLSFLTGSLALSKHVAEATKYFSITVSFGSVKSLISLPCFMSHASIPVE 434
Query: 371 VREVRGLTEDLVRISVGIEDANDLIADLNHAFRTGP 264
VRE RGLTEDLVRISVGIED NDLI DL++A +TGP
Sbjct: 435 VREARGLTEDLVRISVGIEDVNDLIDDLDYALKTGP 470
>gb|AAL36374.1| putative cystathionine beta-lyase precursor CBL [Arabidopsis
thaliana] gi|21281081|gb|AAM45099.1| putative
cystathionine beta-lyase precursor CBL [Arabidopsis
thaliana]
Length = 449
Score = 157 bits (398), Expect = 6e-38
Identities = 78/97 (80%), Positives = 88/97 (90%)
Frame = -2
Query: 551 LHYSQAKGTGSVLSFLTGSLALSKHIVETTKYYSITVSFGSVKSLISLPCFMSHASIPAA 372
LH+SQAKG GSV SF+TGS+ALSKH+VETTKY+SI VSFGSVKSLIS+PCFMSHASIPA
Sbjct: 353 LHFSQAKGAGSVFSFITGSVALSKHLVETTKYFSIAVSFGSVKSLISMPCFMSHASIPAE 412
Query: 371 VREVRGLTEDLVRISVGIEDANDLIADLNHAFRTGPL 261
VRE RGLTEDLVRIS GIED +DLI+DL+ AF+T PL
Sbjct: 413 VREARGLTEDLVRISAGIEDVDDLISDLDIAFKTFPL 449
>ref|NP_191264.1| CYSTATHIONINE BETA-LYASE PRECURSOR (CBL); protein id: At3g57050.1,
supported by cDNA: gi_704396 [Arabidopsis thaliana]
gi|1708993|sp|P53780|METC_ARATH Cystathionine
beta-lyase, chloroplast precursor (CBL)
(Beta-cystathionase) (Cysteine lyase)
gi|2129567|pir||S61429 cystathionine beta-lyase (EC
4.4.1.8) precursor - Arabidopsis thaliana
gi|13786771|pdb|1IBJ|A Chain A, Crystal Structure Of
Cystathionine Beta-Lyase From Arabidopsis Thaliana
gi|13786772|pdb|1IBJ|C Chain C, Crystal Structure Of
Cystathionine Beta-Lyase From Arabidopsis Thaliana
gi|704397|gb|AAA99176.1| cystathionine beta-lyase
gi|6911875|emb|CAB72175.1| CYSTATHIONINE BETA-LYASE
PRECURSOR (CBL) [Arabidopsis thaliana]
Length = 464
Score = 157 bits (398), Expect = 6e-38
Identities = 78/97 (80%), Positives = 88/97 (90%)
Frame = -2
Query: 551 LHYSQAKGTGSVLSFLTGSLALSKHIVETTKYYSITVSFGSVKSLISLPCFMSHASIPAA 372
LH+SQAKG GSV SF+TGS+ALSKH+VETTKY+SI VSFGSVKSLIS+PCFMSHASIPA
Sbjct: 368 LHFSQAKGAGSVFSFITGSVALSKHLVETTKYFSIAVSFGSVKSLISMPCFMSHASIPAE 427
Query: 371 VREVRGLTEDLVRISVGIEDANDLIADLNHAFRTGPL 261
VRE RGLTEDLVRIS GIED +DLI+DL+ AF+T PL
Sbjct: 428 VREARGLTEDLVRISAGIEDVDDLISDLDIAFKTFPL 464
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 449,007,123
Number of Sequences: 1393205
Number of extensions: 8960599
Number of successful extensions: 19702
Number of sequences better than 10.0: 393
Number of HSP's better than 10.0 without gapping: 19358
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19681
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19521267756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)