Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003656A_C01 KMC003656A_c01
(549 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
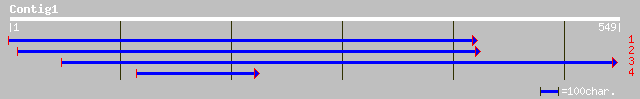
gb|AAK68782.1| putative RING zinc finger ankyrin protein [Arabid... 46 3e-04
ref|NP_180450.1| putative RING zinc finger ankyrin protein; prot... 46 3e-04
gb|AAK58690.1|AF272860_1 receptor-like kinase Xa21-binding prote... 45 6e-04
ref|NP_693378.1| hypothetical protein [Oceanobacillus iheyensis ... 31 8.6
>gb|AAK68782.1| putative RING zinc finger ankyrin protein [Arabidopsis thaliana]
gi|20148259|gb|AAM10020.1| putative RING zinc finger
ankyrin protein [Arabidopsis thaliana]
gi|23397172|gb|AAN31869.1| putative RING zinc finger
ankyrin protein [Arabidopsis thaliana]
Length = 456
Score = 46.2 bits (108), Expect = 3e-04
Identities = 25/39 (64%), Positives = 29/39 (74%), Gaps = 1/39 (2%)
Frame = -3
Query: 478 TKPRRSRKS-NFSEGSSSFKSLSAIGSFGRITGRNSGKI 365
+K R+ R+S N E SSSF LS IGSFGRITGR SG+I
Sbjct: 406 SKFRKHRRSINLGEESSSFMGLSTIGSFGRITGRGSGRI 444
>ref|NP_180450.1| putative RING zinc finger ankyrin protein; protein id: At2g28840.1,
supported by cDNA: gi_13926221 [Arabidopsis thaliana]
gi|25408016|pir||E84689 probable RING zinc finger
ankyrin protein [imported] - Arabidopsis thaliana
gi|3927831|gb|AAC79588.1| putative RING zinc finger
ankyrin protein [Arabidopsis thaliana]
gi|13926222|gb|AAK49587.1|AF370581_1 putative RING zinc
finger ankyrin protein [Arabidopsis thaliana]
Length = 426
Score = 46.2 bits (108), Expect = 3e-04
Identities = 25/39 (64%), Positives = 29/39 (74%), Gaps = 1/39 (2%)
Frame = -3
Query: 478 TKPRRSRKS-NFSEGSSSFKSLSAIGSFGRITGRNSGKI 365
+K R+ R+S N E SSSF LS IGSFGRITGR SG+I
Sbjct: 376 SKFRKHRRSINLGEESSSFMGLSTIGSFGRITGRGSGRI 414
>gb|AAK58690.1|AF272860_1 receptor-like kinase Xa21-binding protein 3 [Oryza sativa]
Length = 450
Score = 45.1 bits (105), Expect = 6e-04
Identities = 28/68 (41%), Positives = 44/68 (64%), Gaps = 3/68 (4%)
Frame = -3
Query: 547 GAILQLIVPTIKNSSDTEVESNP--TKPRRSRKSNFSEGSSSFKSL-SAIGSFGRITGRN 377
G+I +L+V +++ D + S+ T+ R R N SEGSSSFK L SA+GSF ++ GR
Sbjct: 373 GSISRLVVAQTRSACDPDKPSSLQLTRKRSRRSHNLSEGSSSFKGLPSAMGSFSKL-GRG 431
Query: 376 SGKIDDAE 353
S ++ D++
Sbjct: 432 SSRMADSD 439
>ref|NP_693378.1| hypothetical protein [Oceanobacillus iheyensis HTE831]
gi|22778143|dbj|BAC14413.1| hypothetical conserved
protein [Oceanobacillus iheyensis]
Length = 167
Score = 31.2 bits (69), Expect = 8.6
Identities = 14/37 (37%), Positives = 19/37 (50%)
Frame = +1
Query: 397 QMIQWLTSS*SCCYPH*NLIFLISLVWLGWIQLLYHC 507
++ WL C P L FL + +W GWI +LY C
Sbjct: 105 ELTHWL------CIPPAFLFFLWNPIWAGWIMVLYAC 135
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 460,381,526
Number of Sequences: 1393205
Number of extensions: 9460212
Number of successful extensions: 24320
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 23505
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24306
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18947112822
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)