Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003655A_C01 KMC003655A_c01
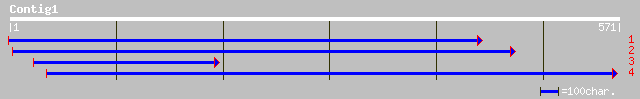
(567 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565124.1| expressed protein; protein id: At1g76010.1, sup... 253 2e-66
emb|CAD41015.1| OSJNBa0042L16.13 [Oryza sativa (japonica cultiva... 244 7e-64
pir||H86335 T20H2.2 protein - Arabidopsis thaliana gi|8778978|gb... 241 6e-63
ref|NP_564108.1| expressed protein; protein id: At1g20220.1, sup... 239 2e-62
gb|AAN60302.1| unknown [Arabidopsis thaliana] 223 2e-57
>ref|NP_565124.1| expressed protein; protein id: At1g76010.1, supported by cDNA:
gi_15724323, supported by cDNA: gi_15809881, supported
by cDNA: gi_16226598 [Arabidopsis thaliana]
gi|15724324|gb|AAL06555.1|AF412102_1 At1g76010/T4O12_22
[Arabidopsis thaliana] gi|15809882|gb|AAL06869.1|
At1g76010/T4O12_22 [Arabidopsis thaliana]
gi|16226599|gb|AAL16210.1|AF428441_1 At1g76010/T4O12_22
[Arabidopsis thaliana] gi|21700865|gb|AAM70556.1|
At1g76010/T4O12_22 [Arabidopsis thaliana]
Length = 350
Score = 253 bits (645), Expect = 2e-66
Identities = 130/161 (80%), Positives = 141/161 (86%), Gaps = 2/161 (1%)
Frame = +2
Query: 89 MDRYQKVEKPRAETPIDENEIRITSQGRMRNYITYAMTLLQEKGSNEIVFKAMGRAINKT 268
MD+YQ+V KP+A+TPID NEIRITSQGR RNYITYAMTLLQ+KGS E+VFKAMGRAINKT
Sbjct: 1 MDKYQRVVKPKADTPIDANEIRITSQGRARNYITYAMTLLQDKGSTEVVFKAMGRAINKT 60
Query: 269 VTIVELIKRRIVGLHQNTAIGSTDITDTWEPLEEGLLPWETTRHVSMITITLSKNELDTA 448
VTIVELIKRRI LHQNT+IGSTDITDTWEP EEGLLP ETTRHVSMITITLSK EL+T+
Sbjct: 61 VTIVELIKRRIPDLHQNTSIGSTDITDTWEPTEEGLLPLETTRHVSMITITLSKIELNTS 120
Query: 449 SVGYQPPLPADQVKAATDFDYEG-ESSPNGRGRGRG-GRGR 565
SVGYQ P+P + VK D DYEG E SP GRGRGRG GRGR
Sbjct: 121 SVGYQCPIPIELVKPMGDIDYEGREGSPGGRGRGRGRGRGR 161
>emb|CAD41015.1| OSJNBa0042L16.13 [Oryza sativa (japonica cultivar-group)]
Length = 262
Score = 244 bits (622), Expect = 7e-64
Identities = 121/159 (76%), Positives = 135/159 (84%)
Frame = +2
Query: 89 MDRYQKVEKPRAETPIDENEIRITSQGRMRNYITYAMTLLQEKGSNEIVFKAMGRAINKT 268
MDRYQ+VEKPR E I NEIRIT+QGR RNYITYA+ LLQ+ ++EIV KAMGRAINKT
Sbjct: 1 MDRYQRVEKPREEAAIGANEIRITAQGRTRNYITYALALLQDNATDEIVIKAMGRAINKT 60
Query: 269 VTIVELIKRRIVGLHQNTAIGSTDITDTWEPLEEGLLPWETTRHVSMITITLSKNELDTA 448
V IVEL+KRRIVGLHQNT+I S DITDTWEPLEEGL ETTRHVS+ITITLSK ELDT+
Sbjct: 61 VAIVELLKRRIVGLHQNTSIESIDITDTWEPLEEGLNTLETTRHVSLITITLSKKELDTS 120
Query: 449 SVGYQPPLPADQVKAATDFDYEGESSPNGRGRGRGGRGR 565
S GYQPP+PADQV+ TDFD E E+ P+GRGRGRG RGR
Sbjct: 121 SPGYQPPIPADQVRPPTDFDQEAEAVPSGRGRGRGRRGR 159
>pir||H86335 T20H2.2 protein - Arabidopsis thaliana
gi|8778978|gb|AAF79893.1|AC022472_2 Contains similarity
to pigpen protein from Mus musculus gb|AF224264 and
contains protein of unknown function DUF78 PF|01918
domain. ESTs gb|N38077, gb|BE037702, gb|AV442191,
gb|AV441368, gb|Z17998, gb|AV527266, gb|AV520794,
gb|AI997847, gb|AV543000 come from this gene.
[Arabidopsis thaliana]
Length = 538
Score = 241 bits (614), Expect = 6e-63
Identities = 125/173 (72%), Positives = 141/173 (81%), Gaps = 3/173 (1%)
Frame = +2
Query: 56 FRKRNFILSIAMDRYQKVEKPRAETPIDENEIRITSQGRMRNYITYAMTLLQEKGSNEIV 235
F + N L MD+YQ+VEKP+A+TPI ENEIRITS GR RNYITYAM LLQE SNE++
Sbjct: 213 FGRSNQNLRNKMDKYQRVEKPKADTPIAENEIRITSMGRARNYITYAMALLQENKSNEVI 272
Query: 236 FKAMGRAINKTVTIVELIKRRIVGLHQNTAIGSTDITDTWEPLEEGLLPWETTRHVSMIT 415
FKAMGRAINK+VTIVELIKRRI GLHQ T+IGSTDITDTWEP EEGL ETTRHVSMIT
Sbjct: 273 FKAMGRAINKSVTIVELIKRRIPGLHQITSIGSTDITDTWEPTEEGLQTIETTRHVSMIT 332
Query: 416 ITLSKNELDTASVGYQPPLPADQVKAATDFDYEG-ESSPNGRG--RGRGGRGR 565
ITLSK +L+T+SVGYQ P+P + VK + DYEG + SP GRG RGRGGRGR
Sbjct: 333 ITLSKEQLNTSSVGYQCPIPIEMVKPLAEIDYEGQDGSPRGRGGRRGRGGRGR 385
>ref|NP_564108.1| expressed protein; protein id: At1g20220.1, supported by cDNA:
gi_16612273 [Arabidopsis thaliana]
gi|16612274|gb|AAL27504.1|AF439833_1 At1g20220/T20H2_3
[Arabidopsis thaliana]
Length = 315
Score = 239 bits (610), Expect = 2e-62
Identities = 122/162 (75%), Positives = 137/162 (84%), Gaps = 3/162 (1%)
Frame = +2
Query: 89 MDRYQKVEKPRAETPIDENEIRITSQGRMRNYITYAMTLLQEKGSNEIVFKAMGRAINKT 268
MD+YQ+VEKP+A+TPI ENEIRITS GR RNYITYAM LLQE SNE++FKAMGRAINK+
Sbjct: 1 MDKYQRVEKPKADTPIAENEIRITSMGRARNYITYAMALLQENKSNEVIFKAMGRAINKS 60
Query: 269 VTIVELIKRRIVGLHQNTAIGSTDITDTWEPLEEGLLPWETTRHVSMITITLSKNELDTA 448
VTIVELIKRRI GLHQ T+IGSTDITDTWEP EEGL ETTRHVSMITITLSK +L+T+
Sbjct: 61 VTIVELIKRRIPGLHQITSIGSTDITDTWEPTEEGLQTIETTRHVSMITITLSKEQLNTS 120
Query: 449 SVGYQPPLPADQVKAATDFDYEG-ESSPNGRG--RGRGGRGR 565
SVGYQ P+P + VK + DYEG + SP GRG RGRGGRGR
Sbjct: 121 SVGYQCPIPIEMVKPLAEIDYEGQDGSPRGRGGRRGRGGRGR 162
>gb|AAN60302.1| unknown [Arabidopsis thaliana]
Length = 272
Score = 223 bits (567), Expect = 2e-57
Identities = 114/162 (70%), Positives = 131/162 (80%), Gaps = 3/162 (1%)
Frame = +2
Query: 89 MDRYQKVEKPRAETPIDENEIRITSQGRMRNYITYAMTLLQEKGSNEIVFKAMGRAINKT 268
MD+YQ+VEKP+A+TPI ENEIRITS GR RNYITYAM LLQE SNE++FKAMGRAINK+
Sbjct: 1 MDKYQRVEKPKADTPIAENEIRITSMGRARNYITYAMALLQENXSNEVIFKAMGRAINKS 60
Query: 269 VTIVELIKRRIVGLHQNTAIGSTDITDTWEPLEEGLLPWETTRHVSMITITLSKNELDTA 448
VTIVELIK+RI GL T+IGSTDITDTWEP EEGL ET +HVS+ TITLSK +T+
Sbjct: 61 VTIVELIKKRIPGLXPITSIGSTDITDTWEPTEEGLQTIETXKHVSIXTITLSKEPXNTS 120
Query: 449 SVGYQPPLPADQVKAATDFDYEG-ESSPNGRG--RGRGGRGR 565
SVGYQ P+P + VK + DYEG + SP GRG RGRGGRGR
Sbjct: 121 SVGYQCPIPIEMVKPLAEXDYEGQDGSPRGRGGRRGRGGRGR 162
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 525,392,612
Number of Sequences: 1393205
Number of extensions: 11829461
Number of successful extensions: 44576
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 40548
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44206
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20669577624
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)