Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003643A_C01 KMC003643A_c01
(571 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
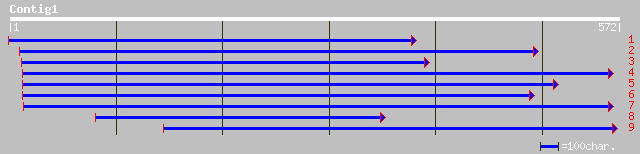
dbj|BAB02395.1| cytochrome P450-like protein [Arabidopsis thaliana] 174 7e-43
ref|NP_188081.1| cytochrome P450, putative; protein id: At3g1463... 174 7e-43
ref|NP_188086.1| cytochrome P450, putative; protein id: At3g1468... 172 2e-42
ref|NP_188079.1| cytochrome P450, putative; protein id: At3g1461... 167 7e-41
ref|NP_188080.1| cytochrome P450, putative; protein id: At3g1462... 167 9e-41
>dbj|BAB02395.1| cytochrome P450-like protein [Arabidopsis thaliana]
Length = 506
Score = 174 bits (441), Expect = 7e-43
Identities = 73/126 (57%), Positives = 100/126 (78%)
Frame = -3
Query: 569 VMFARYLRKDTKLGDLTIPAGVEIIVPVSMLHQEKEFWGDDAMEFNPERFSEGVSKATKG 390
+ R K+ KLGD+T+P G+++ +PV ++H++ + WGDDA EF PERF +G++KATK
Sbjct: 381 IQMNRATHKEIKLGDMTLPGGIQVHMPVLLIHRDTKLWGDDAAEFKPERFKDGIAKATKN 440
Query: 389 KVCYIPFGWGPRLCIGQNFGLLEAKIALSMILQHFSLDLSPSYTHAPSFIITLQPDRGAH 210
+VC++PFGWGPR+CIGQNF LLEAK+AL++ILQ FS +LSPSY H+P + T+ P GAH
Sbjct: 441 QVCFLPFGWGPRICIGQNFALLEAKMALALILQRFSFELSPSYVHSPYRVFTIHPQCGAH 500
Query: 209 LILHKL 192
LILHKL
Sbjct: 501 LILHKL 506
>ref|NP_188081.1| cytochrome P450, putative; protein id: At3g14630.1 [Arabidopsis
thaliana]
Length = 508
Score = 174 bits (441), Expect = 7e-43
Identities = 73/126 (57%), Positives = 100/126 (78%)
Frame = -3
Query: 569 VMFARYLRKDTKLGDLTIPAGVEIIVPVSMLHQEKEFWGDDAMEFNPERFSEGVSKATKG 390
+ R K+ KLGD+T+P G+++ +PV ++H++ + WGDDA EF PERF +G++KATK
Sbjct: 383 IQMNRATHKEIKLGDMTLPGGIQVHMPVLLIHRDTKLWGDDAAEFKPERFKDGIAKATKN 442
Query: 389 KVCYIPFGWGPRLCIGQNFGLLEAKIALSMILQHFSLDLSPSYTHAPSFIITLQPDRGAH 210
+VC++PFGWGPR+CIGQNF LLEAK+AL++ILQ FS +LSPSY H+P + T+ P GAH
Sbjct: 443 QVCFLPFGWGPRICIGQNFALLEAKMALALILQRFSFELSPSYVHSPYRVFTIHPQCGAH 502
Query: 209 LILHKL 192
LILHKL
Sbjct: 503 LILHKL 508
>ref|NP_188086.1| cytochrome P450, putative; protein id: At3g14680.1, supported by
cDNA: gi_13605896 [Arabidopsis thaliana]
gi|9294390|dbj|BAB02400.1| cytochrome P450 [Arabidopsis
thaliana] gi|13605897|gb|AAK32934.1|AF367347_1
AT3g14680/MIE1_18 [Arabidopsis thaliana]
gi|24111277|gb|AAN46762.1| At3g14680/MIE1_18
[Arabidopsis thaliana]
Length = 512
Score = 172 bits (437), Expect = 2e-42
Identities = 79/126 (62%), Positives = 99/126 (77%)
Frame = -3
Query: 569 VMFARYLRKDTKLGDLTIPAGVEIIVPVSMLHQEKEFWGDDAMEFNPERFSEGVSKATKG 390
V R + K+ KLGDLT+P GV+I +PV ++H++ E WG+DA EF PERF +G+SKATK
Sbjct: 387 VQLTRAIHKEMKLGDLTLPGGVQISLPVLLVHRDTELWGNDAGEFKPERFKDGLSKATKN 446
Query: 389 KVCYIPFGWGPRLCIGQNFGLLEAKIALSMILQHFSLDLSPSYTHAPSFIITLQPDRGAH 210
+V + PF WGPR+CIGQNF LLEAK+A+S+ILQ FS +LSPSY HAP IITL P GAH
Sbjct: 447 QVSFFPFAWGPRICIGQNFTLLEAKMAMSLILQRFSFELSPSYVHAPYTIITLYPQFGAH 506
Query: 209 LILHKL 192
L+LHKL
Sbjct: 507 LMLHKL 512
>ref|NP_188079.1| cytochrome P450, putative; protein id: At3g14610.1, supported by
cDNA: gi_18252154 [Arabidopsis thaliana]
gi|9294383|dbj|BAB02393.1| cytochrome P450 [Arabidopsis
thaliana] gi|18252155|gb|AAL61910.1| cytochrome P450
[Arabidopsis thaliana] gi|28059362|gb|AAO30051.1|
cytochrome P450 [Arabidopsis thaliana]
Length = 512
Score = 167 bits (424), Expect = 7e-41
Identities = 72/122 (59%), Positives = 98/122 (80%)
Frame = -3
Query: 557 RYLRKDTKLGDLTIPAGVEIIVPVSMLHQEKEFWGDDAMEFNPERFSEGVSKATKGKVCY 378
R + K+ KLG+LT+PAG++I +P ++ ++ E WGDDA +F PERF +G+SKATK +V +
Sbjct: 391 RVVNKEMKLGELTLPAGIQIYLPTILVQRDTELWGDDAADFKPERFRDGLSKATKNQVSF 450
Query: 377 IPFGWGPRLCIGQNFGLLEAKIALSMILQHFSLDLSPSYTHAPSFIITLQPDRGAHLILH 198
PFGWGPR+CIGQNF +LEAK+A+++ILQ FS +LSPSY HAP ++T +P GAHLILH
Sbjct: 451 FPFGWGPRICIGQNFAMLEAKMAMALILQKFSFELSPSYVHAPQTVMTTRPQFGAHLILH 510
Query: 197 KL 192
KL
Sbjct: 511 KL 512
>ref|NP_188080.1| cytochrome P450, putative; protein id: At3g14620.1, supported by
cDNA: gi_15529168, supported by cDNA: gi_17386113
[Arabidopsis thaliana] gi|9294384|dbj|BAB02394.1|
cytochrome P450 [Arabidopsis thaliana]
gi|15529169|gb|AAK97679.1| AT3g14620/MIE1_12
[Arabidopsis thaliana]
Length = 515
Score = 167 bits (423), Expect = 9e-41
Identities = 73/127 (57%), Positives = 99/127 (77%), Gaps = 1/127 (0%)
Frame = -3
Query: 569 VMFARYLRKDTKLG-DLTIPAGVEIIVPVSMLHQEKEFWGDDAMEFNPERFSEGVSKATK 393
++ R + K+TKLG D+T+P G ++++PV M+H++ E WG+D EFNPERF++G+SKATK
Sbjct: 389 ILLGRTVEKETKLGEDMTLPGGAQVVIPVLMVHRDPELWGEDVHEFNPERFADGISKATK 448
Query: 392 GKVCYIPFGWGPRLCIGQNFGLLEAKIALSMILQHFSLDLSPSYTHAPSFIITLQPDRGA 213
+V ++PFGWGPR C GQNF L+EAK+AL +ILQ FS +LSPSYTHAP ++TL P GA
Sbjct: 449 NQVSFLPFGWGPRFCPGQNFALMEAKMALVLILQRFSFELSPSYTHAPHTVLTLHPQFGA 508
Query: 212 HLILHKL 192
LI H L
Sbjct: 509 PLIFHML 515
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 481,008,825
Number of Sequences: 1393205
Number of extensions: 10169804
Number of successful extensions: 29173
Number of sequences better than 10.0: 2604
Number of HSP's better than 10.0 without gapping: 27590
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28234
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20956655091
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)