Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003635A_C01 KMC003635A_c01
(559 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
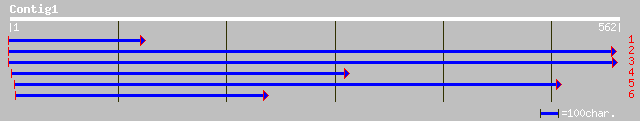
ref|NP_567979.1| putative protein; protein id: At4g35220.1, supp... 169 2e-41
ref|NP_175091.1| hypothetical protein; protein id: At1g44542.1 [... 149 3e-35
ref|NP_567957.1| putative protein; protein id: At4g34180.1, supp... 143 2e-33
ref|NP_782444.1| polyketide cyclase [Clostridium tetani E88] gi|... 73 2e-12
gb|ZP_00056655.1| hypothetical protein [Magnetospirillum magneto... 72 4e-12
>ref|NP_567979.1| putative protein; protein id: At4g35220.1, supported by cDNA:
1368., supported by cDNA: gi_13937203, supported by
cDNA: gi_18491126 [Arabidopsis thaliana]
gi|13937204|gb|AAK50095.1|AF372956_1
AT4g35220/F23E12_220 [Arabidopsis thaliana]
gi|18491127|gb|AAL69532.1| AT4g35220/F23E12_220
[Arabidopsis thaliana] gi|21537400|gb|AAM61741.1|
unknown [Arabidopsis thaliana]
Length = 272
Score = 169 bits (428), Expect = 2e-41
Identities = 81/101 (80%), Positives = 90/101 (88%)
Frame = -2
Query: 558 LNTDRRLMFKKEFDSSYVGFKEDGAKWLVENTDIKLVGVDYLSAASYDHSVPSHLVFLKS 379
LNTDRRLMFKKEFD+SYVGF +DGA+WLV+NTDIKLVG+DYLS A+YD +PSHLVFLK
Sbjct: 171 LNTDRRLMFKKEFDTSYVGFMKDGAQWLVDNTDIKLVGIDYLSVAAYDDLIPSHLVFLKD 230
Query: 378 REIILVEGLKLDDVPAGIYRVQCLPLRLVGSEASPIRCILI 256
RE ILVEGLKLD V AG+Y V CLPLRLVG+E SPIRCILI
Sbjct: 231 RETILVEGLKLDGVKAGLYSVHCLPLRLVGAEGSPIRCILI 271
>ref|NP_175091.1| hypothetical protein; protein id: At1g44542.1 [Arabidopsis
thaliana] gi|13876507|gb|AAK43483.1|AC084807_8
hypothetical protein [Arabidopsis thaliana]
Length = 271
Score = 149 bits (375), Expect = 3e-35
Identities = 72/102 (70%), Positives = 84/102 (81%)
Frame = -2
Query: 558 LNTDRRLMFKKEFDSSYVGFKEDGAKWLVENTDIKLVGVDYLSAASYDHSVPSHLVFLKS 379
LNTDRRLMFKKEFDSS+VGF DGAKWLVENTDIKLVG+DYLS A+YD + +H L+
Sbjct: 170 LNTDRRLMFKKEFDSSFVGFMVDGAKWLVENTDIKLVGLDYLSFAAYDEAPATHRFILER 229
Query: 378 REIILVEGLKLDDVPAGIYRVQCLPLRLVGSEASPIRCILIR 253
R+II VE LKLDDV G+Y + CLPLRLVG+E +P RCILI+
Sbjct: 230 RDIIPVEALKLDDVEVGMYTLHCLPLRLVGAEGAPTRCILIK 271
>ref|NP_567957.1| putative protein; protein id: At4g34180.1, supported by cDNA:
8686., supported by cDNA: gi_14335097, supported by
cDNA: gi_16226602 [Arabidopsis thaliana]
gi|14335098|gb|AAK59828.1| AT4g34180/F28A23_60
[Arabidopsis thaliana]
gi|16226603|gb|AAL16211.1|AF428442_1 AT4g34180/F28A23_60
[Arabidopsis thaliana] gi|21617901|gb|AAM66951.1|
unknown [Arabidopsis thaliana]
gi|21928057|gb|AAM78057.1| AT4g34180/F28A23_60
[Arabidopsis thaliana]
Length = 255
Score = 143 bits (360), Expect = 2e-33
Identities = 68/101 (67%), Positives = 81/101 (79%)
Frame = -2
Query: 555 NTDRRLMFKKEFDSSYVGFKEDGAKWLVENTDIKLVGVDYLSAASYDHSVPSHLVFLKSR 376
NTD+RLMFKKEFDSS+ GF DGAKWLVENTDIKL+G+DYLS A+++ S +H V LK R
Sbjct: 155 NTDKRLMFKKEFDSSFAGFMTDGAKWLVENTDIKLIGLDYLSFAAFEESPATHRVILKGR 214
Query: 375 EIILVEGLKLDDVPAGIYRVQCLPLRLVGSEASPIRCILIR 253
+II VE LKLD V G Y + CLPLRLVG+E +P RCILI+
Sbjct: 215 DIIPVEALKLDGVEVGTYSLHCLPLRLVGAEGAPTRCILIK 255
>ref|NP_782444.1| polyketide cyclase [Clostridium tetani E88]
gi|28203941|gb|AAO36381.1| polyketide cyclase
[Clostridium tetani E88]
Length = 206
Score = 73.2 bits (178), Expect = 2e-12
Identities = 39/91 (42%), Positives = 60/91 (65%)
Frame = -2
Query: 525 EFDSSYVGFKEDGAKWLVENTDIKLVGVDYLSAASYDHSVPSHLVFLKSREIILVEGLKL 346
E++ +V +ED AK+LV+ IK +G+D +S PSH V L+ + I+++E L+L
Sbjct: 117 EYNPKFVYVEEDAAKYLVD-IGIKCIGIDAMSLERDKPHHPSHKVILE-KGIVIIEDLQL 174
Query: 345 DDVPAGIYRVQCLPLRLVGSEASPIRCILIR 253
DV G Y + CLPL++ GSEASP+R +LI+
Sbjct: 175 KDVKEGNYFLSCLPLKIKGSEASPVRAVLIK 205
>gb|ZP_00056655.1| hypothetical protein [Magnetospirillum magnetotacticum]
Length = 214
Score = 72.4 bits (176), Expect = 4e-12
Identities = 37/90 (41%), Positives = 54/90 (59%)
Frame = -2
Query: 522 FDSSYVGFKEDGAKWLVENTDIKLVGVDYLSAASYDHSVPSHLVFLKSREIILVEGLKLD 343
F + ++ F GA+WLVE IKLVG+DYLS + + P+ L I+ VEGL +
Sbjct: 124 FFTDFIAFTPAGAQWLVER-GIKLVGIDYLSVQRFADAEPTTHRVLLGAGIVAVEGLDMR 182
Query: 342 DVPAGIYRVQCLPLRLVGSEASPIRCILIR 253
+ G Y + CLPL+L+G++ SP R +L R
Sbjct: 183 GIDPGEYELVCLPLKLIGADGSPCRVVLTR 212
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 462,037,311
Number of Sequences: 1393205
Number of extensions: 9642588
Number of successful extensions: 21906
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 21421
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21896
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)