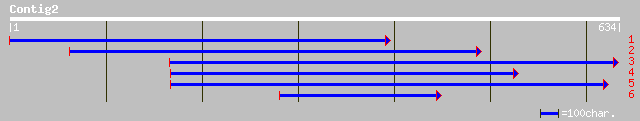
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003634A_C02 KMC003634A_c02
(630 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO72595.1| putative protein kinase [Oryza sativa (japonica c... 169 2e-41
ref|NP_191781.1| serine/threonine protein kinase-like protein; p... 169 4e-41
ref|NP_182229.1| putative protein kinase; protein id: At2g47060.... 167 8e-41
ref|NP_188367.2| putative protein kinase; protein id: At3g17410.... 162 3e-39
dbj|BAB21241.1| Putative Pto kinase interactor 1 [Oryza sativa (... 162 4e-39
>gb|AAO72595.1| putative protein kinase [Oryza sativa (japonica cultivar-group)]
Length = 242
Score = 169 bits (429), Expect = 2e-41
Identities = 83/88 (94%), Positives = 86/88 (97%)
Frame = -2
Query: 629 LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAA 450
LLELLTGRKPVDHTLPRGQQSLVTWATP+LSEDKVRQCVD+RLGG+YPPKAVAK AAVAA
Sbjct: 146 LLELLTGRKPVDHTLPRGQQSLVTWATPRLSEDKVRQCVDSRLGGDYPPKAVAKFAAVAA 205
Query: 449 LCVQYEADFRPNMSIVVKALQPLLTARA 366
LCVQYEADFRPNMSIVVKALQPLL ARA
Sbjct: 206 LCVQYEADFRPNMSIVVKALQPLLNARA 233
>ref|NP_191781.1| serine/threonine protein kinase-like protein; protein id:
At3g62220.1 [Arabidopsis thaliana]
gi|11259788|pir||T48014 serine/threonine protein
kinase-like protein - Arabidopsis thaliana
gi|6899932|emb|CAB71882.1| serine/threonine protein
kinase-like protein [Arabidopsis thaliana]
Length = 361
Score = 169 bits (427), Expect = 4e-41
Identities = 82/87 (94%), Positives = 86/87 (98%)
Frame = -2
Query: 629 LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAA 450
LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV+QCVD+RLGG+YPPKAVAK+AAVAA
Sbjct: 266 LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVKQCVDSRLGGDYPPKAVAKLAAVAA 325
Query: 449 LCVQYEADFRPNMSIVVKALQPLLTAR 369
LCVQYEADFRPNMSIVVKALQPLL AR
Sbjct: 326 LCVQYEADFRPNMSIVVKALQPLLNAR 352
>ref|NP_182229.1| putative protein kinase; protein id: At2g47060.1, supported by
cDNA: gi_13272438, supported by cDNA: gi_19424107,
supported by cDNA: gi_21281065 [Arabidopsis thaliana]
gi|7434311|pir||T02181 probable protein kinase
[imported] - Arabidopsis thaliana
gi|3522961|gb|AAC34243.1| putative protein kinase
[Arabidopsis thaliana]
gi|13272439|gb|AAK17158.1|AF325090_1 putative protein
kinase [Arabidopsis thaliana] gi|19424108|gb|AAL87347.1|
putative protein kinase [Arabidopsis thaliana]
gi|21281066|gb|AAM45092.1| putative protein kinase
[Arabidopsis thaliana]
Length = 365
Score = 167 bits (424), Expect = 8e-41
Identities = 82/88 (93%), Positives = 85/88 (96%)
Frame = -2
Query: 629 LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAA 450
LLELLTGRKPVDH LPRGQQSLVTWATPKLSEDKV+QCVD RLGG+YPPKAVAK+AAVAA
Sbjct: 270 LLELLTGRKPVDHRLPRGQQSLVTWATPKLSEDKVKQCVDARLGGDYPPKAVAKLAAVAA 329
Query: 449 LCVQYEADFRPNMSIVVKALQPLLTARA 366
LCVQYEADFRPNMSIVVKALQPLL ARA
Sbjct: 330 LCVQYEADFRPNMSIVVKALQPLLNARA 357
>ref|NP_188367.2| putative protein kinase; protein id: At3g17410.1, supported by
cDNA: gi_17979366, supported by cDNA: gi_20465524
[Arabidopsis thaliana] gi|11994558|dbj|BAB02745.1|
serine/threonine protein kinase-like protein
[Arabidopsis thaliana] gi|17979367|gb|AAL49909.1|
putative protein kinase [Arabidopsis thaliana]
gi|20465525|gb|AAM20245.1| putative protein kinase
[Arabidopsis thaliana]
Length = 364
Score = 162 bits (411), Expect = 3e-39
Identities = 79/84 (94%), Positives = 82/84 (97%)
Frame = -2
Query: 629 LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAA 450
LLELLTGRKPVDHTLPRGQQS+VTWATPKLSEDKV+QCVD RL GEYPPKAVAK+AAVAA
Sbjct: 266 LLELLTGRKPVDHTLPRGQQSVVTWATPKLSEDKVKQCVDARLNGEYPPKAVAKLAAVAA 325
Query: 449 LCVQYEADFRPNMSIVVKALQPLL 378
LCVQYEADFRPNMSIVVKALQPLL
Sbjct: 326 LCVQYEADFRPNMSIVVKALQPLL 349
>dbj|BAB21241.1| Putative Pto kinase interactor 1 [Oryza sativa (japonica
cultivar-group)] gi|29027802|dbj|BAC65877.1|
OSJNBa0011P19.1 [Oryza sativa (japonica cultivar-group)]
Length = 371
Score = 162 bits (409), Expect = 4e-39
Identities = 79/87 (90%), Positives = 83/87 (94%)
Frame = -2
Query: 629 LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAA 450
LLELLTGRKPVDHTLPRGQQSLVTWATP+LSEDKV+QCVD RL G+YPPKAVAKMAAVAA
Sbjct: 268 LLELLTGRKPVDHTLPRGQQSLVTWATPRLSEDKVKQCVDPRLEGDYPPKAVAKMAAVAA 327
Query: 449 LCVQYEADFRPNMSIVVKALQPLLTAR 369
LCVQYEADFRPNMSIVVKAL PLL +R
Sbjct: 328 LCVQYEADFRPNMSIVVKALNPLLNSR 354
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 503,390,969
Number of Sequences: 1393205
Number of extensions: 10101577
Number of successful extensions: 25744
Number of sequences better than 10.0: 959
Number of HSP's better than 10.0 without gapping: 24313
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25347
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25870486187
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)