Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003586A_C01 KMC003586A_c01
(608 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
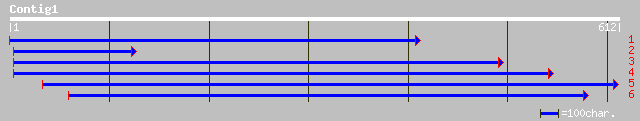
ref|NP_683509.1| DNA repair protein RAD23, putative; protein id:... 206 1e-52
ref|NP_565216.2| DNA repair protein RAD23, putative; protein id:... 206 1e-52
pir||T14337 RAD23 protein, isoform II - carrot gi|1914685|emb|CA... 197 7e-50
ref|NP_173070.1| hypothetical protein; protein id: At1g16190.1 [... 194 1e-48
pir||H86296 F3O9.1 protein - Arabidopsis thaliana gi|4966345|gb|... 180 1e-44
>ref|NP_683509.1| DNA repair protein RAD23, putative; protein id: At1g79650.1,
supported by cDNA: 40579. [Arabidopsis thaliana]
gi|21593616|gb|AAM65583.1| DNA repair protein RAD23,
putative [Arabidopsis thaliana]
Length = 371
Score = 206 bits (525), Expect = 1e-52
Identities = 105/126 (83%), Positives = 113/126 (89%), Gaps = 2/126 (1%)
Frame = -3
Query: 606 FLRNDPQSQALRSMVQSNPQILQPVLQELGKQNPSLLRLIDEHHAEFLQLINEPVEGS-- 433
FLRN+ Q Q LR+MV SNPQILQP+LQELGKQNP LLRLI E+ AEFLQL+NEP EGS
Sbjct: 246 FLRNNDQFQQLRTMVHSNPQILQPMLQELGKQNPQLLRLIQENQAEFLQLVNEPYEGSDG 305
Query: 432 EGDIFDQPEQEMPHAINVTPAEQEAIGRLEAMGFDRASVIEAFLACDRDEQLAANYLLEN 253
EGD+FDQPEQEMPHAINVTPAEQEAI RLEAMGFDRA VIEAFLACDR+E+LAANYLLEN
Sbjct: 306 EGDMFDQPEQEMPHAINVTPAEQEAIQRLEAMGFDRALVIEAFLACDRNEELAANYLLEN 365
Query: 252 PGDFED 235
GDFED
Sbjct: 366 SGDFED 371
>ref|NP_565216.2| DNA repair protein RAD23, putative; protein id: At1g79650.2,
supported by cDNA: gi_14334441, supported by cDNA:
gi_17104776 [Arabidopsis thaliana]
gi|14334442|gb|AAK59419.1| putative DNA repair protein
RAD23 [Arabidopsis thaliana] gi|17104777|gb|AAL34277.1|
putative DNA repair protein RAD23 [Arabidopsis thaliana]
Length = 365
Score = 206 bits (525), Expect = 1e-52
Identities = 105/126 (83%), Positives = 113/126 (89%), Gaps = 2/126 (1%)
Frame = -3
Query: 606 FLRNDPQSQALRSMVQSNPQILQPVLQELGKQNPSLLRLIDEHHAEFLQLINEPVEGS-- 433
FLRN+ Q Q LR+MV SNPQILQP+LQELGKQNP LLRLI E+ AEFLQL+NEP EGS
Sbjct: 240 FLRNNDQFQQLRTMVHSNPQILQPMLQELGKQNPQLLRLIQENQAEFLQLVNEPYEGSDG 299
Query: 432 EGDIFDQPEQEMPHAINVTPAEQEAIGRLEAMGFDRASVIEAFLACDRDEQLAANYLLEN 253
EGD+FDQPEQEMPHAINVTPAEQEAI RLEAMGFDRA VIEAFLACDR+E+LAANYLLEN
Sbjct: 300 EGDMFDQPEQEMPHAINVTPAEQEAIQRLEAMGFDRALVIEAFLACDRNEELAANYLLEN 359
Query: 252 PGDFED 235
GDFED
Sbjct: 360 SGDFED 365
>pir||T14337 RAD23 protein, isoform II - carrot gi|1914685|emb|CAA72742.1| RAD23
protein, isoform II [Daucus carota]
Length = 379
Score = 197 bits (502), Expect = 7e-50
Identities = 98/124 (79%), Positives = 106/124 (85%)
Frame = -3
Query: 606 FLRNDPQSQALRSMVQSNPQILQPVLQELGKQNPSLLRLIDEHHAEFLQLINEPVEGSEG 427
FLRN+PQ Q LRSMVQ NPQILQP+L ELGKQNP LLR I EHH EFLQLINEPVE SEG
Sbjct: 256 FLRNNPQFQTLRSMVQRNPQILQPMLLELGKQNPQLLRQIQEHHEEFLQLINEPVEASEG 315
Query: 426 DIFDQPEQEMPHAINVTPAEQEAIGRLEAMGFDRASVIEAFLACDRDEQLAANYLLENPG 247
D+FDQPEQ++P I VT A+QEAI RLEAMGFDR VIEAFLACDR+E+LA NYLLEN G
Sbjct: 316 DMFDQPEQDVPQEITVTAADQEAIERLEAMGFDRGLVIEAFLACDRNEELAVNYLLENAG 375
Query: 246 DFED 235
DFED
Sbjct: 376 DFED 379
>ref|NP_173070.1| hypothetical protein; protein id: At1g16190.1 [Arabidopsis
thaliana]
Length = 368
Score = 194 bits (492), Expect = 1e-48
Identities = 97/126 (76%), Positives = 110/126 (86%), Gaps = 2/126 (1%)
Frame = -3
Query: 606 FLRNDPQSQALRSMVQSNPQILQPVLQELGKQNPSLLRLIDEHHAEFLQLINEPVEGSEG 427
FLR + Q Q LRSMV SNPQILQP+LQELGKQNP LLRLI E+ AEFLQL+NEP EGS+G
Sbjct: 243 FLRGNDQFQQLRSMVNSNPQILQPMLQELGKQNPQLLRLIQENQAEFLQLLNEPYEGSDG 302
Query: 426 D--IFDQPEQEMPHAINVTPAEQEAIGRLEAMGFDRASVIEAFLACDRDEQLAANYLLEN 253
D IFDQP+QEMPH++NVTP EQE+I RLEAMGFDRA VIEAFL+CDR+E+LAANYLLE+
Sbjct: 303 DVDIFDQPDQEMPHSVNVTPEEQESIERLEAMGFDRAIVIEAFLSCDRNEELAANYLLEH 362
Query: 252 PGDFED 235
DFED
Sbjct: 363 SADFED 368
>pir||H86296 F3O9.1 protein - Arabidopsis thaliana
gi|4966345|gb|AAD34676.1|AC006341_4 Similar to gb|Y12014
RAD23 protein isoform II from Daucus carota. This gene
is probably cut off. EST gb|AA651284 comes from this
gene. [Arabidopsis thaliana]
Length = 113
Score = 180 bits (457), Expect = 1e-44
Identities = 89/113 (78%), Positives = 101/113 (88%), Gaps = 2/113 (1%)
Frame = -3
Query: 567 MVQSNPQILQPVLQELGKQNPSLLRLIDEHHAEFLQLINEPVEGSEGD--IFDQPEQEMP 394
MV SNPQILQP+LQELGKQNP LLRLI E+ AEFLQL+NEP EGS+GD IFDQP+QEMP
Sbjct: 1 MVNSNPQILQPMLQELGKQNPQLLRLIQENQAEFLQLLNEPYEGSDGDVDIFDQPDQEMP 60
Query: 393 HAINVTPAEQEAIGRLEAMGFDRASVIEAFLACDRDEQLAANYLLENPGDFED 235
H++NVTP EQE+I RLEAMGFDRA VIEAFL+CDR+E+LAANYLLE+ DFED
Sbjct: 61 HSVNVTPEEQESIERLEAMGFDRAIVIEAFLSCDRNEELAANYLLEHSADFED 113
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 517,861,661
Number of Sequences: 1393205
Number of extensions: 11200814
Number of successful extensions: 35279
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 33304
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34928
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24283162270
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)