Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003584A_C01 KMC003584A_c01
(802 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
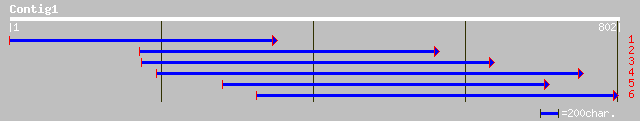
ref|NP_565843.1| expressed protein; protein id: At2g36310.1, sup... 201 8e-51
pir||B84779 hypothetical protein At2g36310 [imported] - Arabidop... 129 7e-29
ref|NP_563745.1| expressed protein; protein id: At1g05620.1, sup... 87 3e-16
pir||C86190 hypothetical protein [imported] - Arabidopsis thalia... 87 3e-16
gb|EAA02229.1| agCP13080 [Anopheles gambiae str. PEST] 60 3e-08
>ref|NP_565843.1| expressed protein; protein id: At2g36310.1, supported by cDNA:
2576., supported by cDNA: gi_15809829, supported by
cDNA: gi_17978870 [Arabidopsis thaliana]
gi|15809830|gb|AAL06843.1| At2g36310/F2H17.8
[Arabidopsis thaliana] gi|17978871|gb|AAL47407.1|
At2g36310/F2H17.8 [Arabidopsis thaliana]
gi|20197937|gb|AAD21435.2| expressed protein
[Arabidopsis thaliana] gi|21554564|gb|AAM63615.1|
unknown [Arabidopsis thaliana]
Length = 336
Score = 201 bits (512), Expect = 8e-51
Identities = 89/107 (83%), Positives = 99/107 (92%)
Frame = -1
Query: 793 KGKHVSLLSDMCKFYRDWHVKSDGLHGIFLHDPVSFVAVVRPELFTYKKGVVRVETQGIC 614
KGKH L+SDMCKFYRDWHVKSDG++G++LHDPVSFVAVVRP+LFTYKKGVVRVETQGIC
Sbjct: 230 KGKHSKLISDMCKFYRDWHVKSDGVYGVYLHDPVSFVAVVRPDLFTYKKGVVRVETQGIC 289
Query: 613 VGHTLMDQGLKKWNTSNPWTDYSPVSVAWTVDIEGVLNYVREVLMKP 473
VGHTLMDQGLK+WN SNPW YSP+SVAWTVD+EGVL YV+ LMKP
Sbjct: 290 VGHTLMDQGLKRWNGSNPWVGYSPISVAWTVDVEGVLEYVKAKLMKP 336
>pir||B84779 hypothetical protein At2g36310 [imported] - Arabidopsis thaliana
Length = 305
Score = 129 bits (323), Expect = 7e-29
Identities = 64/102 (62%), Positives = 72/102 (69%), Gaps = 27/102 (26%)
Frame = -1
Query: 793 KGKHVSLLSDMCKFYRDWHVKSDGLHG---------------------------IFLHDP 695
KGKH L+SDMCKFYRDWHVKSDG++G ++LHDP
Sbjct: 195 KGKHSKLISDMCKFYRDWHVKSDGVYGKQESVINQTCLEIAVLILKSLLLLVSGVYLHDP 254
Query: 694 VSFVAVVRPELFTYKKGVVRVETQGICVGHTLMDQGLKKWNT 569
VSFVAVVRP+LFTYKKGVVRVETQGICVGHTLMDQGLK++ T
Sbjct: 255 VSFVAVVRPDLFTYKKGVVRVETQGICVGHTLMDQGLKRFIT 296
>ref|NP_563745.1| expressed protein; protein id: At1g05620.1, supported by cDNA:
39571. [Arabidopsis thaliana] gi|21593497|gb|AAM65464.1|
unknown [Arabidopsis thaliana]
gi|25083300|gb|AAN72060.1| expressed protein
[Arabidopsis thaliana]
Length = 322
Score = 87.0 bits (214), Expect = 3e-16
Identities = 43/107 (40%), Positives = 63/107 (58%)
Frame = -1
Query: 796 SKGKHVSLLSDMCKFYRDWHVKSDGLHGIFLHDPVSFVAVVRPELFTYKKGVVRVETQGI 617
SKGK L + Y D+H+ + + G++LHDP + +A P LFTY +GV RV+T GI
Sbjct: 215 SKGKLAQYLCKILDVYYDYHLTAYEIKGVYLHDPATILAAFLPSLFTYTEGVARVQTSGI 274
Query: 616 CVGHTLMDQGLKKWNTSNPWTDYSPVSVAWTVDIEGVLNYVREVLMK 476
G TL+ LK++ +N W+D V VA TVD V+ + + LM+
Sbjct: 275 TRGLTLLYNNLKRFEEANEWSDKPTVKVAVTVDAPAVVKLIMDRLME 321
>pir||C86190 hypothetical protein [imported] - Arabidopsis thaliana
gi|4836912|gb|AAD30614.1|AC007153_6 Hypothetical protein
[Arabidopsis thaliana]
Length = 358
Score = 87.0 bits (214), Expect = 3e-16
Identities = 43/107 (40%), Positives = 63/107 (58%)
Frame = -1
Query: 796 SKGKHVSLLSDMCKFYRDWHVKSDGLHGIFLHDPVSFVAVVRPELFTYKKGVVRVETQGI 617
SKGK L + Y D+H+ + + G++LHDP + +A P LFTY +GV RV+T GI
Sbjct: 251 SKGKLAQYLCKILDVYYDYHLTAYEIKGVYLHDPATILAAFLPSLFTYTEGVARVQTSGI 310
Query: 616 CVGHTLMDQGLKKWNTSNPWTDYSPVSVAWTVDIEGVLNYVREVLMK 476
G TL+ LK++ +N W+D V VA TVD V+ + + LM+
Sbjct: 311 TRGLTLLYNNLKRFEEANEWSDKPTVKVAVTVDAPAVVKLIMDRLME 357
>gb|EAA02229.1| agCP13080 [Anopheles gambiae str. PEST]
Length = 356
Score = 60.5 bits (145), Expect = 3e-08
Identities = 35/108 (32%), Positives = 55/108 (50%)
Frame = -1
Query: 802 KESKGKHVSLLSDMCKFYRDWHVKSDGLHGIFLHDPVSFVAVVRPELFTYKKGVVRVETQ 623
++ G+ L D+ +FY ++ + GL G +HDP + V+ P LFT ++G VRV +
Sbjct: 244 RDDAGEPGRFLWDVSRFYLRFYSERIGLDGCHVHDPSAIAYVIDPTLFTLREGPVRVINE 303
Query: 622 GICVGHTLMDQGLKKWNTSNPWTDYSPVSVAWTVDIEGVLNYVREVLM 479
G +GHTL K + + W DY V V + +L RE L+
Sbjct: 304 GPAIGHTLQKFDGKS-HPHDGWADYRAQQVGVAVRDQALLALYRETLV 350
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 713,600,866
Number of Sequences: 1393205
Number of extensions: 15993870
Number of successful extensions: 33823
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 32643
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33805
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40616159090
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)