Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003581A_C02 KMC003581A_c02
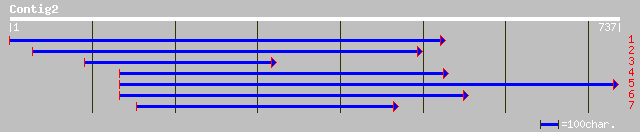
(697 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAA85874.1| orf UL135 38 0.12
dbj|BAB29221.1| unnamed protein product [Mus musculus] 38 0.12
ref|XP_133029.1| RIKEN cDNA 3110069A13 [Mus musculus] 38 0.12
pir||PQ0452 extensin-like protein - Persian tobacco (strain S2S3... 37 0.20
ref|NP_608462.1| CG14619-PA [Drosophila melanogaster] gi|2464378... 37 0.34
>gb|AAA85874.1| orf UL135
Length = 328
Score = 38.1 bits (87), Expect = 0.12
Identities = 35/112 (31%), Positives = 47/112 (41%), Gaps = 2/112 (1%)
Frame = +3
Query: 351 PQFPINPEQNISYSSSSSILF*SKSFTFMSDSNSYSLNQPVIPGLRWMEMKKMRRKISET 530
P P + ++ S+ SSSS L S S + S P PG R+K T
Sbjct: 141 PGTPTSIGRSPSHCSSSSSLSSSTSVDTVLYQPPPSWKPPPPPG---------RKKRPPT 191
Query: 531 QKKRTHK*KTRSAQPSTEVP--NKGLKQPP*QALPPPSPTRPPPISGGPPST 680
R + S +P T +P K L PP + PP PT+P P+ PP T
Sbjct: 192 PPVRAPTTRLSSHRPPTPIPAPRKNLSTPPTKKTPP--PTKPKPVGWTPPVT 241
>dbj|BAB29221.1| unnamed protein product [Mus musculus]
Length = 475
Score = 38.1 bits (87), Expect = 0.12
Identities = 22/57 (38%), Positives = 34/57 (59%)
Frame = +3
Query: 486 RWMEMKKMRRKISETQKKRTHK*KTRSAQPSTEVPNKGLKQPP*QALPPPSPTRPPP 656
R ++KK RK+ ++ T +T + +PS++ P + L+QPP LPP PT PPP
Sbjct: 411 RNFDLKKHVRKLHDSSLGLT---RTPTGEPSSDPPPQ-LQQPPPAPLPPLQPTLPPP 463
>ref|XP_133029.1| RIKEN cDNA 3110069A13 [Mus musculus]
Length = 475
Score = 38.1 bits (87), Expect = 0.12
Identities = 22/57 (38%), Positives = 34/57 (59%)
Frame = +3
Query: 486 RWMEMKKMRRKISETQKKRTHK*KTRSAQPSTEVPNKGLKQPP*QALPPPSPTRPPP 656
R ++KK RK+ ++ T +T + +PS++ P + L+QPP LPP PT PPP
Sbjct: 411 RNFDLKKHVRKLHDSSLGLT---RTPTGEPSSDPPPQ-LQQPPPAPLPPLQPTLPPP 463
>pir||PQ0452 extensin-like protein - Persian tobacco (strain S2S3) (fragment)
Length = 122
Score = 37.4 bits (85), Expect = 0.20
Identities = 19/45 (42%), Positives = 25/45 (55%), Gaps = 2/45 (4%)
Frame = +3
Query: 558 TRSAQPSTEVPNKG--LKQPP*QALPPPSPTRPPPISGGPPSTRS 686
+RS P E P+ +K PP + PPPSP+ PPP PP + S
Sbjct: 34 SRSPPPKREQPSPPPPVKSPPPPSPPPPSPSPPPPSPSPPPPSPS 78
Score = 32.7 bits (73), Expect = 4.9
Identities = 15/37 (40%), Positives = 18/37 (48%)
Frame = +3
Query: 564 SAQPSTEVPNKGLKQPP*QALPPPSPTRPPPISGGPP 674
S P + P PP + PPPSP+ PPP PP
Sbjct: 45 SPPPPVKSPPPPSPPPPSPSPPPPSPSPPPPSPSPPP 81
>ref|NP_608462.1| CG14619-PA [Drosophila melanogaster] gi|24643789|ref|NP_728453.1|
CG14619-PD [Drosophila melanogaster]
gi|24643791|ref|NP_728454.1| CG14619-PE [Drosophila
melanogaster] gi|17862942|gb|AAL39948.1| SD04280p
[Drosophila melanogaster] gi|22832690|gb|AAF50952.2|
CG14619-PA [Drosophila melanogaster]
gi|22832691|gb|AAN09564.1| CG14619-PD [Drosophila
melanogaster] gi|22832692|gb|AAN09565.1| CG14619-PE
[Drosophila melanogaster]
Length = 856
Score = 36.6 bits (83), Expect = 0.34
Identities = 19/53 (35%), Positives = 25/53 (46%)
Frame = +3
Query: 531 QKKRTHK*KTRSAQPSTEVPNKGLKQPP*QALPPPSPTRPPPISGGPPSTRSG 689
+ KRT + + ST V G PP PPP P PPP +G P+ +G
Sbjct: 351 RSKRTRRNQAEEVPTSTGVEQAGPAAPP--PTPPPPPPPPPPTNGHKPAESNG 401
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 667,405,084
Number of Sequences: 1393205
Number of extensions: 16122096
Number of successful extensions: 98924
Number of sequences better than 10.0: 141
Number of HSP's better than 10.0 without gapping: 60692
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 88436
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 31684559424
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)