Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
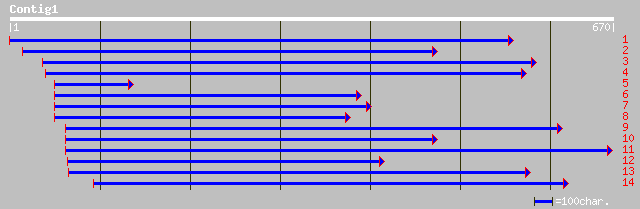
Query= KMC003578A_C01 KMC003578A_c01
(648 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL27697.1| 5-enolpyruvylshikimate-3-phosphate synthase [Dicl... 236 3e-61
gb|AAL27698.1|AF371966_1 5-enolpyruvylshikimate-3-phosphate synt... 236 3e-61
sp|P23281|ARO2_TOBAC 3-phosphoshikimate 1-carboxyvinyltransferas... 235 3e-61
sp|P11043|AROA_PETHY 3-phosphoshikimate 1-carboxyvinyltransferas... 234 7e-61
pir||XUTOVS 3-phosphoshikimate 1-carboxyvinyltransferase (EC 2.5... 231 5e-60
>gb|AAL27697.1| 5-enolpyruvylshikimate-3-phosphate synthase [Dicliptera chinensis]
Length = 516
Score = 236 bits (601), Expect = 3e-61
Identities = 112/124 (90%), Positives = 120/124 (96%)
Frame = -2
Query: 647 LQGIDVNMNKMPDVAMTLAVVALFANGPTAIRDVASWRVKETERMIAICTELRKLGATVE 468
L+ IDVNMNKMPDVAMTLAVVALFA+GPTAIRDVASWRVKETERMIAICTELRKLGATVE
Sbjct: 391 LRAIDVNMNKMPDVAMTLAVVALFADGPTAIRDVASWRVKETERMIAICTELRKLGATVE 450
Query: 467 EGPDYCVITPPEKLNITSIDTYDDHRMAMAFSLAACGDVPVTIKDPGCTRKTFPDYFEVL 288
EGPDYC+ITPPEKLN+T+IDTYDDHRMAMAFSLAAC DVPVTIKDPGCTRKTFP+YF+VL
Sbjct: 451 EGPDYCIITPPEKLNVTAIDTYDDHRMAMAFSLAACADVPVTIKDPGCTRKTFPNYFDVL 510
Query: 287 ERFS 276
+S
Sbjct: 511 STYS 514
>gb|AAL27698.1|AF371966_1 5-enolpyruvylshikimate-3-phosphate synthase B [Dicliptera
chinensis]
Length = 516
Score = 236 bits (601), Expect = 3e-61
Identities = 112/124 (90%), Positives = 120/124 (96%)
Frame = -2
Query: 647 LQGIDVNMNKMPDVAMTLAVVALFANGPTAIRDVASWRVKETERMIAICTELRKLGATVE 468
L+ IDVNMNKMPDVAMTLAVVALFA+GPTAIRDVASWRVKETERMIAICTELRKLGATVE
Sbjct: 391 LRAIDVNMNKMPDVAMTLAVVALFADGPTAIRDVASWRVKETERMIAICTELRKLGATVE 450
Query: 467 EGPDYCVITPPEKLNITSIDTYDDHRMAMAFSLAACGDVPVTIKDPGCTRKTFPDYFEVL 288
EGPDYC+ITPPEKLN+T+IDTYDDHRMAMAFSLAAC DVPVTIKDPGCTRKTFP+YF+VL
Sbjct: 451 EGPDYCIITPPEKLNVTAIDTYDDHRMAMAFSLAACADVPVTIKDPGCTRKTFPNYFDVL 510
Query: 287 ERFS 276
+S
Sbjct: 511 STYS 514
>sp|P23281|ARO2_TOBAC 3-phosphoshikimate 1-carboxyvinyltransferase 2
(5-enolpyruvylshikimate-3-phosphate synthase 2) (EPSP
synthase 2) gi|100297|pir||S18354 3-phosphoshikimate
1-carboxyvinyltransferase (EC 2.5.1.19) - common tobacco
gi|170231|gb|AAA34072.1|
5-enolpyruvylshikimate-3-phosphate synthase
Length = 338
Score = 235 bits (600), Expect = 3e-61
Identities = 111/125 (88%), Positives = 121/125 (96%)
Frame = -2
Query: 647 LQGIDVNMNKMPDVAMTLAVVALFANGPTAIRDVASWRVKETERMIAICTELRKLGATVE 468
L+ IDVNMNKMPDVAMTLAVVALFA+GPTAIRDVASWRVKETERMIAICTELRKLGATVE
Sbjct: 213 LRAIDVNMNKMPDVAMTLAVVALFADGPTAIRDVASWRVKETERMIAICTELRKLGATVE 272
Query: 467 EGPDYCVITPPEKLNITSIDTYDDHRMAMAFSLAACGDVPVTIKDPGCTRKTFPDYFEVL 288
EGPDYC+ITPPEKLN+T IDTYDDHRMAMAFSLAAC DVPVTI DPGCTRKTFP+YF+VL
Sbjct: 273 EGPDYCIITPPEKLNVTEIDTYDDHRMAMAFSLAACADVPVTINDPGCTRKTFPNYFDVL 332
Query: 287 ERFSQ 273
+++S+
Sbjct: 333 QQYSK 337
>sp|P11043|AROA_PETHY 3-phosphoshikimate 1-carboxyvinyltransferase, chloroplast precursor
(5-enolpyruvylshikimate-3-phosphate synthase) (EPSP
synthase) gi|66620|pir||XUPJVS 3-phosphoshikimate
1-carboxyvinyltransferase (EC 2.5.1.19) precursor -
garden petunia gi|169191|gb|AAA33699.1|
5-enolpyruvylshikimate-3-phosphate synthase precursor
Length = 516
Score = 234 bits (597), Expect = 7e-61
Identities = 110/125 (88%), Positives = 121/125 (96%)
Frame = -2
Query: 647 LQGIDVNMNKMPDVAMTLAVVALFANGPTAIRDVASWRVKETERMIAICTELRKLGATVE 468
L+ IDVNMNKMPDVAMTLAVVAL+A+GPTAIRDVASWRVKETERMIAICTELRKLGATVE
Sbjct: 391 LRAIDVNMNKMPDVAMTLAVVALYADGPTAIRDVASWRVKETERMIAICTELRKLGATVE 450
Query: 467 EGPDYCVITPPEKLNITSIDTYDDHRMAMAFSLAACGDVPVTIKDPGCTRKTFPDYFEVL 288
EGPDYC+ITPPEKLN+T IDTYDDHRMAMAFSLAAC DVPVTI DPGCTRKTFP+YF+VL
Sbjct: 451 EGPDYCIITPPEKLNVTDIDTYDDHRMAMAFSLAACADVPVTINDPGCTRKTFPNYFDVL 510
Query: 287 ERFSQ 273
+++S+
Sbjct: 511 QQYSK 515
>pir||XUTOVS 3-phosphoshikimate 1-carboxyvinyltransferase (EC 2.5.1.19)
precursor - tomato
Length = 520
Score = 231 bits (590), Expect = 5e-60
Identities = 110/125 (88%), Positives = 119/125 (95%)
Frame = -2
Query: 647 LQGIDVNMNKMPDVAMTLAVVALFANGPTAIRDVASWRVKETERMIAICTELRKLGATVE 468
L+ IDVNMNKMPDVAMTLAVVALFA+GPT IRDVASWRVKETERMIAICTELRKLGATV
Sbjct: 395 LRAIDVNMNKMPDVAMTLAVVALFADGPTTIRDVASWRVKETERMIAICTELRKLGATVV 454
Query: 467 EGPDYCVITPPEKLNITSIDTYDDHRMAMAFSLAACGDVPVTIKDPGCTRKTFPDYFEVL 288
EG DYC+ITPPEKLN+T IDTYDDHRMAMAFSLAAC DVPVTIK+PGCTRKTFPDYFEVL
Sbjct: 455 EGSDYCIITPPEKLNVTEIDTYDDHRMAMAFSLAACADVPVTIKNPGCTRKTFPDYFEVL 514
Query: 287 ERFSQ 273
+++S+
Sbjct: 515 QKYSK 519
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 522,240,484
Number of Sequences: 1393205
Number of extensions: 10623915
Number of successful extensions: 44588
Number of sequences better than 10.0: 229
Number of HSP's better than 10.0 without gapping: 42062
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44218
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27576232529
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)