Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003572A_C01 KMC003572A_c01
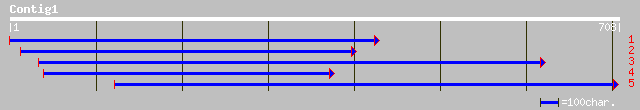
(702 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_180470.1| putative SCARECROW gene regulator; protein id: ... 158 8e-38
ref|NP_172232.1| transcription factor scarecrow-like 14, putativ... 156 3e-37
gb|AAF79548.1|AC022464_6 F22G5.9 [Arabidopsis thaliana] 156 3e-37
dbj|BAB86447.1| P0406G08.7 [Oryza sativa (japonica cultivar-group)] 149 5e-35
ref|NP_172233.1| transcription factor scarecrow-like 14, putativ... 141 8e-33
>ref|NP_180470.1| putative SCARECROW gene regulator; protein id: At2g29060.1
[Arabidopsis thaliana] gi|7487873|pir||T02736 probable
SCARECROW gene regulator [imported] - Arabidopsis
thaliana gi|3461846|gb|AAC33232.1| putative SCARECROW
gene regulator [Arabidopsis thaliana]
Length = 1336
Score = 158 bits (399), Expect = 8e-38
Identities = 74/112 (66%), Positives = 87/112 (77%), Gaps = 1/112 (0%)
Frame = -1
Query: 702 ATFDMFDXVISRDNQWRRLIERESVGREAMNVIACEGLERVERPETYKRWQVRNTRAGFK 523
A FDMFD + RDN+ R ERE GREAMNVIACE +RVERPETY++WQVR RAGFK
Sbjct: 1225 ALFDMFDSTLPRDNKERIRFEREFYGREAMNVIACEEADRVERPETYRQWQVRMVRAGFK 1284
Query: 522 QLPLNEGLMDKFRTKLKKW-YHKDFVFDEDNNWMLQGWKGRIMYASTCWVPA 370
Q + L++ FR KLKKW YHKDFV DE++ W+LQGWKGR +YAS+CWVPA
Sbjct: 1285 QKTIKPELVELFRGKLKKWRYHKDFVVDENSKWLLQGWKGRTLYASSCWVPA 1336
Score = 128 bits (322), Expect = 7e-29
Identities = 58/109 (53%), Positives = 80/109 (73%), Gaps = 1/109 (0%)
Frame = -1
Query: 696 FDMFDXVISRDNQWRRLIERESVGREAMNVIACEGLERVERPETYKRWQVRNTRAGFKQL 517
FDM D ++R++ R + E+E GRE MNV+ACEG ERVERPE+YK+WQ R RAGF+Q+
Sbjct: 585 FDMCDTNLTREDPMRVMFEKEFYGREIMNVVACEGTERVERPESYKQWQARAMRAGFRQI 644
Query: 516 PLNEGLMDKFRTKLKKWYH-KDFVFDEDNNWMLQGWKGRIMYASTCWVP 373
PL + L+ K + ++ Y K+F D+D +W+LQGWKGRI+Y S+ WVP
Sbjct: 645 PLEKELVQKLKLMVESGYKPKEFDVDQDCHWLLQGWKGRIVYGSSIWVP 693
>ref|NP_172232.1| transcription factor scarecrow-like 14, putative; protein id:
At1g07520.1 [Arabidopsis thaliana]
Length = 695
Score = 156 bits (394), Expect = 3e-37
Identities = 71/112 (63%), Positives = 87/112 (77%), Gaps = 1/112 (0%)
Frame = -1
Query: 702 ATFDMFDXVISRDNQWRRLIERESVGREAMNVIACEGLERVERPETYKRWQVRNTRAGFK 523
A FD+F +S++N R E E GRE MNVIACEG++RVERPETYK+WQVR RAGFK
Sbjct: 584 ALFDLFGATLSKENPERIHFEGEFYGREVMNVIACEGVDRVERPETYKQWQVRMIRAGFK 643
Query: 522 QLPLNEGLMDKFRTKLKKW-YHKDFVFDEDNNWMLQGWKGRIMYASTCWVPA 370
Q P+ L+ FR K+KKW YHKDFV DED+NW LQGWKGRI+++S+CWVP+
Sbjct: 644 QKPVEAELVQLFREKMKKWGYHKDFVLDEDSNWFLQGWKGRILFSSSCWVPS 695
>gb|AAF79548.1|AC022464_6 F22G5.9 [Arabidopsis thaliana]
Length = 1502
Score = 156 bits (394), Expect = 3e-37
Identities = 71/112 (63%), Positives = 87/112 (77%), Gaps = 1/112 (0%)
Frame = -1
Query: 702 ATFDMFDXVISRDNQWRRLIERESVGREAMNVIACEGLERVERPETYKRWQVRNTRAGFK 523
A FD+F +S++N R E E GRE MNVIACEG++RVERPETYK+WQVR RAGFK
Sbjct: 1391 ALFDLFGATLSKENPERIHFEGEFYGREVMNVIACEGVDRVERPETYKQWQVRMIRAGFK 1450
Query: 522 QLPLNEGLMDKFRTKLKKW-YHKDFVFDEDNNWMLQGWKGRIMYASTCWVPA 370
Q P+ L+ FR K+KKW YHKDFV DED+NW LQGWKGRI+++S+CWVP+
Sbjct: 1451 QKPVEAELVQLFREKMKKWGYHKDFVLDEDSNWFLQGWKGRILFSSSCWVPS 1502
Score = 129 bits (323), Expect = 5e-29
Identities = 57/107 (53%), Positives = 77/107 (71%)
Frame = -1
Query: 702 ATFDMFDXVISRDNQWRRLIERESVGREAMNVIACEGLERVERPETYKRWQVRNTRAGFK 523
A FDM D ++R+++ R + E+E GRE +NV+ACEG ERVERPETYK+WQ R RAGF+
Sbjct: 657 AVFDMCDSKLAREDEMRLMYEKEFYGREIVNVVACEGTERVERPETYKQWQARLIRAGFR 716
Query: 522 QLPLNEGLMDKFRTKLKKWYHKDFVFDEDNNWMLQGWKGRIMYASTC 382
QLPL + LM + K++ Y K+F D++ NW+LQGWKGRI+ C
Sbjct: 717 QLPLEKELMQNLKLKIENGYDKNFDVDQNGNWLLQGWKGRIVCKQCC 763
>dbj|BAB86447.1| P0406G08.7 [Oryza sativa (japonica cultivar-group)]
Length = 818
Score = 149 bits (375), Expect = 5e-35
Identities = 68/110 (61%), Positives = 85/110 (76%)
Frame = -1
Query: 702 ATFDMFDXVISRDNQWRRLIERESVGREAMNVIACEGLERVERPETYKRWQVRNTRAGFK 523
A FDM + I +DN+ R LIE REA+NVI+CEGLER+ERPETYK+WQVRN R GFK
Sbjct: 704 AIFDMLETNIPKDNEQRLLIESALFSREAINVISCEGLERMERPETYKQWQVRNQRVGFK 763
Query: 522 QLPLNEGLMDKFRTKLKKWYHKDFVFDEDNNWMLQGWKGRIMYASTCWVP 373
QLPLN+ +M + R K+ + YHKDF+ DEDN W+LQGWKGRI++A + W P
Sbjct: 764 QLPLNQDMMKRAREKV-RCYHKDFIIDEDNRWLLQGWKGRILFALSTWKP 812
>ref|NP_172233.1| transcription factor scarecrow-like 14, putative; protein id:
At1g07530.1 [Arabidopsis thaliana]
Length = 769
Score = 141 bits (356), Expect = 8e-33
Identities = 62/111 (55%), Positives = 84/111 (74%)
Frame = -1
Query: 702 ATFDMFDXVISRDNQWRRLIERESVGREAMNVIACEGLERVERPETYKRWQVRNTRAGFK 523
A FDM D ++R+++ R + E+E GRE +NV+ACEG ERVERPETYK+WQ R RAGF+
Sbjct: 657 AVFDMCDSKLAREDEMRLMYEKEFYGREIVNVVACEGTERVERPETYKQWQARLIRAGFR 716
Query: 522 QLPLNEGLMDKFRTKLKKWYHKDFVFDEDNNWMLQGWKGRIMYASTCWVPA 370
QLPL + LM + K++ Y K+F D++ NW+LQGWKGRI+YAS+ WVP+
Sbjct: 717 QLPLEKELMQNLKLKIENGYDKNFDVDQNGNWLLQGWKGRIVYASSLWVPS 767
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 572,608,249
Number of Sequences: 1393205
Number of extensions: 12366648
Number of successful extensions: 28956
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 27815
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28898
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 32250355128
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)