Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003566A_C02 KMC003566A_c02
(667 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
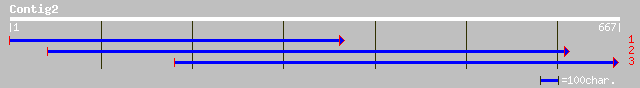
ref|NP_524112.1| survival motor neuron CG16725-PA gi|7294092|gb|... 37 0.31
pir||JE0291 FB19 protein - human gi|2117159|emb|CAA73697.1| FB19... 35 1.2
ref|NP_002705.2| protein phosphatase 1, regulatory subunit 10; p... 35 1.2
gb|EAA37030.1| GLP_16_10672_15699 [Giardia lamblia ATCC 50803] 33 2.6
ref|XP_148784.2| RIKEN cDNA 2610025H06 [Mus musculus] 33 3.5
>ref|NP_524112.1| survival motor neuron CG16725-PA gi|7294092|gb|AAF49446.1|
CG16725-PA [Drosophila melanogaster]
gi|10444398|gb|AAG17893.1|AF296281_1 survival motor
neuron protein SMN [Drosophila melanogaster]
gi|16197975|gb|AAL13758.1| LD23602p [Drosophila
melanogaster]
Length = 226
Score = 36.6 bits (83), Expect = 0.31
Identities = 23/71 (32%), Positives = 36/71 (50%), Gaps = 3/71 (4%)
Frame = -3
Query: 611 EDEKIRKMAMGA---AERVLSAVRTNISSDLNVNEEKERKDSELEQIGGSETDLTAVLNA 441
EDE++ + A ++ S+ R+ IS L + + Q G+E D A+L A
Sbjct: 138 EDEQLSRPKASAGSHSKTPKSSRRSRISGGLVMPPMPPVPPMIVGQGDGAEQDFVAMLTA 197
Query: 440 WYSAGFYTGKY 408
WY +G+YTG Y
Sbjct: 198 WYMSGYYTGLY 208
>pir||JE0291 FB19 protein - human gi|2117159|emb|CAA73697.1| FB19 protein [Homo
sapiens]
Length = 940
Score = 34.7 bits (78), Expect = 1.2
Identities = 20/68 (29%), Positives = 36/68 (52%)
Frame = -3
Query: 653 NNSVELDKKMLFPCEDEKIRKMAMGAAERVLSAVRTNISSDLNVNEEKERKDSELEQIGG 474
NN+ +L K++ EDE++RK+A ++ +R+ S+ ++K+RKD +
Sbjct: 111 NNTAKLVKQLSKSSEDEELRKLASVLVSDWMAVIRSQSSTQPAEKDKKKRKDEGKSRTTL 170
Query: 473 SETDLTAV 450
E LT V
Sbjct: 171 PERPLTEV 178
>ref|NP_002705.2| protein phosphatase 1, regulatory subunit 10; phosphatase nuclear
targeting subunit [Homo sapiens]
gi|15277231|dbj|BAB63324.1| FB19 [Homo sapiens]
gi|27544390|dbj|BAC54929.1| protein phosphatase 1,
regulatory subunit 10 [Homo sapiens]
gi|28411188|emb|CAD67521.1| protein phospahatase nuclear
targeting subunit [Homo sapiens]
Length = 940
Score = 34.7 bits (78), Expect = 1.2
Identities = 20/68 (29%), Positives = 36/68 (52%)
Frame = -3
Query: 653 NNSVELDKKMLFPCEDEKIRKMAMGAAERVLSAVRTNISSDLNVNEEKERKDSELEQIGG 474
NN+ +L K++ EDE++RK+A ++ +R+ S+ ++K+RKD +
Sbjct: 111 NNTAKLVKQLSKSSEDEELRKLASVLVSDWMAVIRSQSSTQPAEKDKKKRKDEGKSRTTL 170
Query: 473 SETDLTAV 450
E LT V
Sbjct: 171 PERPLTEV 178
>gb|EAA37030.1| GLP_16_10672_15699 [Giardia lamblia ATCC 50803]
Length = 1675
Score = 33.5 bits (75), Expect = 2.6
Identities = 17/35 (48%), Positives = 25/35 (70%)
Frame = -3
Query: 575 AERVLSAVRTNISSDLNVNEEKERKDSELEQIGGS 471
AE V +A+R ++ NV+EE ERKD+EL+Q+ S
Sbjct: 1199 AESVSNALRAAGNAGKNVSEELERKDAELQQLRAS 1233
>ref|XP_148784.2| RIKEN cDNA 2610025H06 [Mus musculus]
Length = 863
Score = 33.1 bits (74), Expect = 3.5
Identities = 19/68 (27%), Positives = 36/68 (52%)
Frame = -3
Query: 653 NNSVELDKKMLFPCEDEKIRKMAMGAAERVLSAVRTNISSDLNVNEEKERKDSELEQIGG 474
NN+ +L K++ EDE++RK+A ++ +R+ S+ ++K+RK+ +
Sbjct: 111 NNTAKLVKQLSKSSEDEELRKLASVLVSDWMAVIRSQSSTQPAEKDKKKRKEEGKSRTTL 170
Query: 473 SETDLTAV 450
E LT V
Sbjct: 171 PERPLTEV 178
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 522,328,095
Number of Sequences: 1393205
Number of extensions: 10234304
Number of successful extensions: 26549
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 25679
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26538
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28855580904
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)