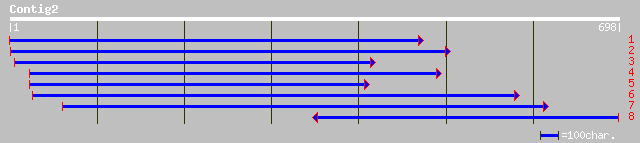
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003546A_C02 KMC003546A_c02
(697 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_197343.1| putative protein; protein id: At5g18420.1 [Arab... 259 2e-68
gb|AAM53304.1| putative protein [Arabidopsis thaliana] 258 7e-68
ref|NP_060016.1| hypothetical protein C40 [Homo sapiens] gi|6048... 252 3e-66
dbj|BAC30912.1| unnamed protein product [Mus musculus] 252 3e-66
gb|AAH18664.1| Unknown (protein for MGC:2536) [Homo sapiens] 252 3e-66
>ref|NP_197343.1| putative protein; protein id: At5g18420.1 [Arabidopsis thaliana]
Length = 441
Score = 259 bits (663), Expect = 2e-68
Identities = 130/168 (77%), Positives = 146/168 (86%), Gaps = 1/168 (0%)
Frame = -2
Query: 696 AAVRDLIAKALKGALAPTQQEQVLLELGNDPKLVYHCGLTPRKLPELVENNPLIAVDVLT 517
A VRDL+ K LK L+PT+QE ++ EL NDPKLV+HCG+TPRKLP+LVE+NP IAV++LT
Sbjct: 265 ATVRDLLVKGLKVTLSPTEQEDIITELANDPKLVFHCGITPRKLPQLVEHNPQIAVEILT 324
Query: 516 KLINSPEIAEYFTVLVNMDMSLHSMEVVNRLTTAVELPSEFIHMYITNCISSCVSIK-DK 340
KL NS EI +Y+ L NMDMSLHSMEVVNRLTTAVELP +FI MYITNCISSC + K DK
Sbjct: 325 KLNNSTEINDYYEALGNMDMSLHSMEVVNRLTTAVELPKDFIRMYITNCISSCENAKQDK 384
Query: 339 YMQNRLVRLVCVFLQSLIRNNIINVKDLFIEVQAFCIEFSRIREAAAL 196
YMQNRLVRLVCVFLQSLIRN IINVKDLFIEVQAFCIEFSR+REAA L
Sbjct: 385 YMQNRLVRLVCVFLQSLIRNKIINVKDLFIEVQAFCIEFSRVREAAGL 432
>gb|AAM53304.1| putative protein [Arabidopsis thaliana]
Length = 441
Score = 258 bits (658), Expect = 7e-68
Identities = 129/168 (76%), Positives = 146/168 (86%), Gaps = 1/168 (0%)
Frame = -2
Query: 696 AAVRDLIAKALKGALAPTQQEQVLLELGNDPKLVYHCGLTPRKLPELVENNPLIAVDVLT 517
A VRDL+ K LK L+PT+QE ++ EL NDPKLV+HCG+TPR+LP+LVE+NP IAV++LT
Sbjct: 265 ATVRDLLVKGLKVTLSPTEQEDIVTELANDPKLVFHCGITPRELPQLVEHNPQIAVEILT 324
Query: 516 KLINSPEIAEYFTVLVNMDMSLHSMEVVNRLTTAVELPSEFIHMYITNCISSCVSIK-DK 340
KL NS EI +Y+ L NMDMSLHSMEVVNRLTTAVELP +FI MYITNCISSC + K DK
Sbjct: 325 KLNNSTEINDYYEALGNMDMSLHSMEVVNRLTTAVELPKDFIRMYITNCISSCENAKQDK 384
Query: 339 YMQNRLVRLVCVFLQSLIRNNIINVKDLFIEVQAFCIEFSRIREAAAL 196
YMQNRLVRLVCVFLQSLIRN IINVKDLFIEVQAFCIEFSR+REAA L
Sbjct: 385 YMQNRLVRLVCVFLQSLIRNKIINVKDLFIEVQAFCIEFSRVREAAGL 432
>ref|NP_060016.1| hypothetical protein C40 [Homo sapiens]
gi|6048964|gb|AAF02418.1|AF103798_1 unknown [Homo
sapiens]
Length = 510
Score = 252 bits (644), Expect = 3e-66
Identities = 125/165 (75%), Positives = 144/165 (86%)
Frame = -2
Query: 690 VRDLIAKALKGALAPTQQEQVLLELGNDPKLVYHCGLTPRKLPELVENNPLIAVDVLTKL 511
++ ++AKA K L+ QQ Q+L EL DPKLVYH GLTP KLP+LVENNPL+A+++L KL
Sbjct: 326 IKRIMAKAFKSPLSSPQQTQLLGELEKDPKLVYHIGLTPAKLPDLVENNPLVAIEMLLKL 385
Query: 510 INSPEIAEYFTVLVNMDMSLHSMEVVNRLTTAVELPSEFIHMYITNCISSCVSIKDKYMQ 331
+ S +I EYF+VLVNMDMSLHSMEVVNRLTTAV+LP EFIH+YI+NCIS+C IKDKYMQ
Sbjct: 386 MQSSQITEYFSVLVNMDMSLHSMEVVNRLTTAVDLPPEFIHLYISNCISTCEQIKDKYMQ 445
Query: 330 NRLVRLVCVFLQSLIRNNIINVKDLFIEVQAFCIEFSRIREAAAL 196
NRLVRLVCVFLQSLIRN IINV+DLFIEVQAFCIEFSRIREAA L
Sbjct: 446 NRLVRLVCVFLQSLIRNKIINVQDLFIEVQAFCIEFSRIREAAGL 490
>dbj|BAC30912.1| unnamed protein product [Mus musculus]
Length = 292
Score = 252 bits (644), Expect = 3e-66
Identities = 125/165 (75%), Positives = 144/165 (86%)
Frame = -2
Query: 690 VRDLIAKALKGALAPTQQEQVLLELGNDPKLVYHCGLTPRKLPELVENNPLIAVDVLTKL 511
++ ++AKA K L+ QQ Q+L EL DPKLVYH GLTP KLP+LVENNPL+A+++L KL
Sbjct: 108 IKRIMAKAFKSPLSSPQQTQLLGELEKDPKLVYHIGLTPAKLPDLVENNPLVAIEMLLKL 167
Query: 510 INSPEIAEYFTVLVNMDMSLHSMEVVNRLTTAVELPSEFIHMYITNCISSCVSIKDKYMQ 331
+ S +I EYF+VLVNMDMSLHSMEVVNRLTTAV+LP EFIH+YI+NCIS+C IKDKYMQ
Sbjct: 168 MQSSQITEYFSVLVNMDMSLHSMEVVNRLTTAVDLPPEFIHLYISNCISTCEQIKDKYMQ 227
Query: 330 NRLVRLVCVFLQSLIRNNIINVKDLFIEVQAFCIEFSRIREAAAL 196
NRLVRLVCVFLQSLIRN IINV+DLFIEVQAFCIEFSRIREAA L
Sbjct: 228 NRLVRLVCVFLQSLIRNKIINVQDLFIEVQAFCIEFSRIREAAGL 272
>gb|AAH18664.1| Unknown (protein for MGC:2536) [Homo sapiens]
Length = 450
Score = 252 bits (644), Expect = 3e-66
Identities = 125/165 (75%), Positives = 144/165 (86%)
Frame = -2
Query: 690 VRDLIAKALKGALAPTQQEQVLLELGNDPKLVYHCGLTPRKLPELVENNPLIAVDVLTKL 511
++ ++AKA K L+ QQ Q+L EL DPKLVYH GLTP KLP+LVENNPL+A+++L KL
Sbjct: 266 IKRIMAKAFKSPLSSPQQTQLLGELEKDPKLVYHIGLTPAKLPDLVENNPLVAIEMLLKL 325
Query: 510 INSPEIAEYFTVLVNMDMSLHSMEVVNRLTTAVELPSEFIHMYITNCISSCVSIKDKYMQ 331
+ S +I EYF+VLVNMDMSLHSMEVVNRLTTAV+LP EFIH+YI+NCIS+C IKDKYMQ
Sbjct: 326 MQSSQITEYFSVLVNMDMSLHSMEVVNRLTTAVDLPPEFIHLYISNCISTCEQIKDKYMQ 385
Query: 330 NRLVRLVCVFLQSLIRNNIINVKDLFIEVQAFCIEFSRIREAAAL 196
NRLVRLVCVFLQSLIRN IINV+DLFIEVQAFCIEFSRIREAA L
Sbjct: 386 NRLVRLVCVFLQSLIRNKIINVQDLFIEVQAFCIEFSRIREAAGL 430
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 511,957,017
Number of Sequences: 1393205
Number of extensions: 10149783
Number of successful extensions: 23940
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 22925
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23917
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 31684559424
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)