Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003545A_C01 KMC003545A_c01
(618 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
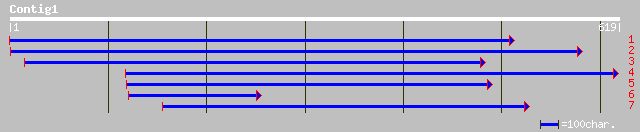
ref|NP_444406.1| histidine-rich glycoprotein [Mus musculus] gi|1... 36 0.46
dbj|BAB33095.1| histidine-rich glycoprotein [Mus musculus] gi|15... 36 0.46
gb|AAG28416.1|AF194028_1 histidine-rich glycoprotein [Mus musculus] 36 0.46
ref|NP_064483.1| multidomain presynaptic cytomatrix protein Picc... 35 1.0
gb|AAF07822.2|AF138789_1 multidomain presynaptic cytomatrix prot... 35 1.0
>ref|NP_444406.1| histidine-rich glycoprotein [Mus musculus]
gi|13358880|dbj|BAB33094.1| histidine-rich glycoprotein
[Mus musculus]
Length = 525
Score = 35.8 bits (81), Expect = 0.46
Identities = 19/65 (29%), Positives = 27/65 (41%)
Frame = -3
Query: 580 QETGKAENSLPVHFQQAIPVYAAPPTRIDEPSLIQKEELPTISTPGQQDKLPPRVQETEN 401
++ G + P H QQ VY PP I E + + P+ S P L P +Q
Sbjct: 439 RKRGPGKGLFPFHHQQIGYVYRLPPLNIGEVLTLPEANFPSFSLPNCNRSLQPEIQPFPQ 498
Query: 400 SPSES 386
+ S S
Sbjct: 499 TASRS 503
>dbj|BAB33095.1| histidine-rich glycoprotein [Mus musculus]
gi|15029888|gb|AAH11168.1| Unknown (protein for
MGC:19088) [Mus musculus]
Length = 525
Score = 35.8 bits (81), Expect = 0.46
Identities = 19/65 (29%), Positives = 27/65 (41%)
Frame = -3
Query: 580 QETGKAENSLPVHFQQAIPVYAAPPTRIDEPSLIQKEELPTISTPGQQDKLPPRVQETEN 401
++ G + P H QQ VY PP I E + + P+ S P L P +Q
Sbjct: 439 RKRGPGKGLFPFHHQQIGYVYRLPPLNIGEVLTLPEANFPSFSLPNCNRSLQPEIQPFPQ 498
Query: 400 SPSES 386
+ S S
Sbjct: 499 TASRS 503
>gb|AAG28416.1|AF194028_1 histidine-rich glycoprotein [Mus musculus]
Length = 525
Score = 35.8 bits (81), Expect = 0.46
Identities = 19/65 (29%), Positives = 27/65 (41%)
Frame = -3
Query: 580 QETGKAENSLPVHFQQAIPVYAAPPTRIDEPSLIQKEELPTISTPGQQDKLPPRVQETEN 401
++ G + P H QQ VY PP I E + + P+ S P L P +Q
Sbjct: 439 RKRGPGKGLFPFHHQQIGYVYRLPPLNIGEVLTLPEANFPSFSLPNCNRSLQPEIQPFPQ 498
Query: 400 SPSES 386
+ S S
Sbjct: 499 TASRS 503
>ref|NP_064483.1| multidomain presynaptic cytomatrix protein Piccolo; piccolo
(presynaptic cytomatrix protein) [Rattus norvegicus]
gi|24212076|sp|Q9JKS6|PCLO_RAT Piccolo protein
(Multidomain presynaptic cytomatrix protein)
gi|7528227|gb|AAF63196.1| multidomain presynaptic
cytomatrix protein Piccolo [Rattus norvegicus]
Length = 5085
Score = 34.7 bits (78), Expect = 1.0
Identities = 23/89 (25%), Positives = 43/89 (47%), Gaps = 3/89 (3%)
Frame = -3
Query: 616 IPCQQDILPHKIQETGKAENSLPVHFQQAIPVYAAPPTRIDEPSLIQKEELPTISTPGQQ 437
IP + ++ E + + + + Q +P + PP + PS+ +K+EL + P Q
Sbjct: 33 IPAGMEADLSQLSEEERRQIAAVMSRAQGLPKGSVPPAAAESPSMHRKQELDSSQAPQQP 92
Query: 436 DKLPPRVQETENSPSES---ETVRNLEKL 359
K P + T+ S+S +T R+ +KL
Sbjct: 93 GKPPDPGRPTQPGLSKSRTTDTFRSEQKL 121
>gb|AAF07822.2|AF138789_1 multidomain presynaptic cytomatrix protein Piccolo [Rattus
norvegicus]
Length = 4880
Score = 34.7 bits (78), Expect = 1.0
Identities = 23/89 (25%), Positives = 43/89 (47%), Gaps = 3/89 (3%)
Frame = -3
Query: 616 IPCQQDILPHKIQETGKAENSLPVHFQQAIPVYAAPPTRIDEPSLIQKEELPTISTPGQQ 437
IP + ++ E + + + + Q +P + PP + PS+ +K+EL + P Q
Sbjct: 33 IPAGMEADLSQLSEEERRQIAAVMSRAQGLPKGSVPPAAAESPSMHRKQELDSSQAPQQP 92
Query: 436 DKLPPRVQETENSPSES---ETVRNLEKL 359
K P + T+ S+S +T R+ +KL
Sbjct: 93 GKPPDPGRPTQPGLSKSRTTDTFRSEQKL 121
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 487,838,416
Number of Sequences: 1393205
Number of extensions: 9791590
Number of successful extensions: 23223
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 22265
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23204
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 24733321959
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)