Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003528A_C01 KMC003528A_c01
(1146 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
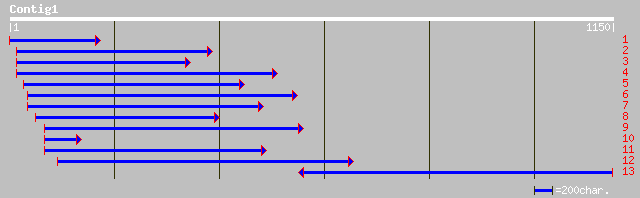
emb|CAC09928.1| hypothetical protein [Catharanthus roseus] 157 2e-37
ref|NP_565660.1| expressed protein; protein id: At2g27830.1, sup... 150 3e-35
ref|NP_194007.1| similar to selenium-binding protein-like; prote... 63 8e-09
dbj|BAB33412.1| putative senescence-associated protein [Pisum sa... 61 4e-08
ref|NP_565000.1| expressed protein; protein id: At1g70780.1, sup... 54 6e-06
>emb|CAC09928.1| hypothetical protein [Catharanthus roseus]
Length = 196
Score = 157 bits (398), Expect = 2e-37
Identities = 97/203 (47%), Positives = 130/203 (63%), Gaps = 11/203 (5%)
Frame = -2
Query: 872 SKNHRSCHV--ETHRRGKLAERSSSFCSQTSTSTSSSSSAASMPAAKLRRPKTVPDLKLP 699
++NHR E +R+G+LAE++ SF + TS ++ + L RP+TVP+L L
Sbjct: 6 NQNHRKSGSVGEKNRKGRLAEKALSFHAGTSVGNAAETML-------LNRPRTVPNL-LS 57
Query: 698 AGADV---SPR---QPPKL---LLKVTMMRSLGPVLMVMSPESTVGDLVAATLRQYVKEG 546
G V SP QPPKL LL VT+M S+GP+ ++ SPE+TV DL++A LRQY KEG
Sbjct: 58 GGKFVDKTSPEFQFQPPKLTKLLLNVTIMGSVGPIQVLTSPEATVEDLISAALRQYSKEG 117
Query: 545 RRPILPSGDPSQFDLHYSQFSLESLDRKEKLSDIGSRNFFMCPRGDVSGKDSNVTTPFAA 366
RRPIL S D + FDLHYSQFSLESL R+EKL +GSRNFF+CP+ + T ++
Sbjct: 118 RRPILKSTDAAGFDLHYSQFSLESLHREEKLISLGSRNFFLCPK-----RCGAEPTASSS 172
Query: 365 SCSREVDQDGGSGGGWLKFMDFL 297
CS+E D+ +G WLKFM+ L
Sbjct: 173 GCSKEADKATKTGTPWLKFMELL 195
>ref|NP_565660.1| expressed protein; protein id: At2g27830.1, supported by cDNA:
33700., supported by cDNA: gi_17473712 [Arabidopsis
thaliana] gi|25407927|pir||E84677 hypothetical protein
At2g27830 [imported] - Arabidopsis thaliana
gi|3860249|gb|AAC73017.1| expressed protein [Arabidopsis
thaliana] gi|17473713|gb|AAL38309.1| unknown protein
[Arabidopsis thaliana] gi|21386999|gb|AAM47903.1|
unknown protein [Arabidopsis thaliana]
gi|21592892|gb|AAM64842.1| unknown [Arabidopsis
thaliana]
Length = 190
Score = 150 bits (380), Expect = 3e-35
Identities = 89/189 (47%), Positives = 120/189 (63%), Gaps = 7/189 (3%)
Frame = -2
Query: 845 ETHRRGKLAERSSSFCSQTSTSTSSSSSAASMPAAKLRRPKTVPDL-------KLPAGAD 687
++HRR KL+E++ SF + +T S+ +LRRPKT+P+L +P
Sbjct: 14 KSHRR-KLSEKAMSFHGRGTTPLSNPG--------ELRRPKTLPELFSTGQSITVPETVS 64
Query: 686 VSPRQPPKLLLKVTMMRSLGPVLMVMSPESTVGDLVAATLRQYVKEGRRPILPSGDPSQF 507
+ PR KLLL VT+ SLG V +++SPESTV DL+ A +RQYVKE RRP LP +PS+F
Sbjct: 65 LPPRLT-KLLLNVTVQGSLGAVQIIISPESTVSDLIDAAVRQYVKEARRPFLPESEPSRF 123
Query: 506 DLHYSQFSLESLDRKEKLSDIGSRNFFMCPRGDVSGKDSNVTTPFAASCSREVDQDGGSG 327
DLHYSQFSLES+ R EKL +GSRNFF+C R + G + + SCS+E ++ +G
Sbjct: 124 DLHYSQFSLESIVRDEKLISLGSRNFFLCGRKETGGFIGCGLS--SESCSKEAEKVAKTG 181
Query: 326 GGWLKFMDF 300
WLKFM F
Sbjct: 182 FHWLKFMGF 190
>ref|NP_194007.1| similar to selenium-binding protein-like; protein id: At4g22760.1
[Arabidopsis thaliana] gi|7486881|pir||T04571
hypothetical protein T12H17.150 - Arabidopsis thaliana
gi|2827553|emb|CAA16561.1| predicted protein
[Arabidopsis thaliana] gi|7269122|emb|CAB79231.1|
predicted protein [Arabidopsis thaliana]
Length = 889
Score = 63.2 bits (152), Expect = 8e-09
Identities = 32/106 (30%), Positives = 59/106 (55%)
Frame = -2
Query: 716 PDLKLPAGADVSPRQPPKLLLKVTMMRSLGPVLMVMSPESTVGDLVAATLRQYVKEGRRP 537
P P + + ++ PK+++ V + S GPV ++ V + + + +Y KEGR P
Sbjct: 106 PSSSSPINQEGNTKEAPKVIISVAVEGSPGPVRAMVKLSCNVEETIKIVVDKYCKEGRTP 165
Query: 536 ILPSGDPSQFDLHYSQFSLESLDRKEKLSDIGSRNFFMCPRGDVSG 399
L S F+LH S FS++ L+++E + ++GSR+F+M + +G
Sbjct: 166 KLDRD--SAFELHQSHFSIQCLEKREIIGELGSRSFYMRKKAPETG 209
>dbj|BAB33412.1| putative senescence-associated protein [Pisum sativum]
Length = 221
Score = 60.8 bits (146), Expect = 4e-08
Identities = 36/118 (30%), Positives = 58/118 (48%), Gaps = 2/118 (1%)
Frame = -2
Query: 665 KLLLKVTMMRSLGPVLMVMSPESTVGDLVAATLRQYVKEGRRPILPSGDPSQFDLHYSQF 486
+LL+ + + S GP+ V+ E V ++ L+ Y +EGR P+L GD F L+
Sbjct: 53 RLLISINVFGSAGPIRFVVKEEELVAAVIDTALKSYAREGRLPVL-GGDIRTFALYCPLV 111
Query: 485 SLESLDRKEKLSDIGSRNFFMC--PRGDVSGKDSNVTTPFAASCSREVDQDGGSGGGW 318
++L + + +G+RNF +C P+ + +N T S SR GGSG W
Sbjct: 112 GSDALSPWDTIGSVGARNFMLCKKPQATTGDEAANGTGTEVVSRSR-----GGSGKAW 164
>ref|NP_565000.1| expressed protein; protein id: At1g70780.1, supported by cDNA:
gi_13358235, supported by cDNA: gi_15146213, supported
by cDNA: gi_16226337 [Arabidopsis thaliana]
gi|25372855|pir||D96732 hypothetical protein F15H11.3
[imported] - Arabidopsis thaliana
gi|5902391|gb|AAD55493.1|AC008148_3 Unknown protein
[Arabidopsis thaliana]
gi|11762244|gb|AAG40399.1|AF325047_1 At1g70780
[Arabidopsis thaliana]
gi|12324748|gb|AAG52330.1|AC011663_9 unknown protein;
8615-9128 [Arabidopsis thaliana]
gi|15146214|gb|AAK83590.1| At1g70780/F5A18_4
[Arabidopsis thaliana] gi|23506215|gb|AAN31119.1|
At1g70780/F5A18_4 [Arabidopsis thaliana]
Length = 140
Score = 53.5 bits (127), Expect = 6e-06
Identities = 33/118 (27%), Positives = 62/118 (51%), Gaps = 1/118 (0%)
Frame = -2
Query: 665 KLLLKVTMMRSLGPVLMVMSPESTVGDLVAATLRQYVKEGRRPILPSGDPSQFDLHYSQF 486
++L+ VT++ S GP+ V + V ++ L+ Y +EGR P+L S D + F L+
Sbjct: 17 RILISVTVLGSAGPIRFVAYEDDLVASVIDTALKGYAREGRLPLLGS-DFNDFLLYCPMV 75
Query: 485 SLESLDRKEKLSDIGSRNFFMCPRGDVSG-KDSNVTTPFAASCSREVDQDGGSGGGWL 315
E+L + + +G+RNF +C + + ++SN + + +R+ GGS W+
Sbjct: 76 GPEALSTWDAIGSLGARNFMLCRKPEEKKVEESNGRSDSTINGARK----GGSLKAWI 129
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,076,730,813
Number of Sequences: 1393205
Number of extensions: 27628367
Number of successful extensions: 186981
Number of sequences better than 10.0: 660
Number of HSP's better than 10.0 without gapping: 111292
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 167927
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 70281887232
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)