Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
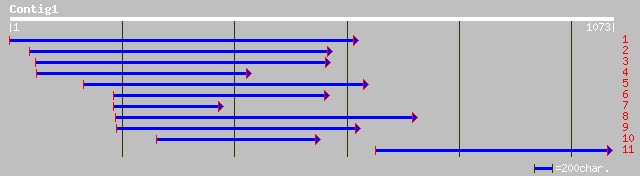
Query= KMC003525A_C01 KMC003525A_c01
(1071 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565138.1| expressed protein; protein id: At1g76670.1, sup... 332 5e-90
dbj|BAC42299.1| unknown protein [Arabidopsis thaliana] 330 2e-89
ref|NP_564133.1| expressed protein; protein id: At1g21070.1, sup... 330 2e-89
gb|AAM61035.1| unknown [Arabidopsis thaliana] 328 9e-89
ref|NP_199057.1| putative protein; protein id: At5g42420.1 [Arab... 313 3e-84
>ref|NP_565138.1| expressed protein; protein id: At1g76670.1, supported by cDNA:
gi_14532697, supported by cDNA: gi_16604379, supported by
cDNA: gi_18491194 [Arabidopsis thaliana]
gi|25354481|pir||A96795 unknown protein F28O16.4
[imported] - Arabidopsis thaliana
gi|6143887|gb|AAF04433.1|AC010718_2 unknown protein;
11341-9662 [Arabidopsis thaliana]
gi|14532698|gb|AAK64150.1| unknown protein [Arabidopsis
thaliana] gi|16604380|gb|AAL24196.1| At1g76670/F28O16_4
[Arabidopsis thaliana] gi|18491195|gb|AAL69500.1| unknown
protein [Arabidopsis thaliana] gi|23308311|gb|AAN18125.1|
At1g76670/F28O16_4 [Arabidopsis thaliana]
Length = 347
Score = 332 bits (852), Expect = 5e-90
Identities = 159/204 (77%), Positives = 182/204 (88%)
Frame = -3
Query: 1069 GVGVCTVTDVKVNLKGFVCACLAILSTSLQQISIGSLQKKYSIGSFELLSKTAPIQAVSL 890
GVG+CTVTDVKVN KGF+CAC A+ STSLQQISIGSLQKKYS+GSFELLSKTAPIQA+SL
Sbjct: 143 GVGICTVTDVKVNAKGFICACTAVFSTSLQQISIGSLQKKYSVGSFELLSKTAPIQAISL 202
Query: 889 LVLGPFIDYYLSAKLITNYKMSSGAILFILLSCTLAVFCNVSQYLCIGRFSAVSFQVLGH 710
L+ GPF+DY LS K I+ Y+M+ GAI ILLSC LAVFCN+SQYLCIGRFSA SFQVLGH
Sbjct: 203 LICGPFVDYLLSGKFISTYQMTYGAIFCILLSCALAVFCNISQYLCIGRFSATSFQVLGH 262
Query: 709 MKTVCVLTLGWLLFDSELTFKNIMGMVLAVVGMVIYSWAVEVEKQSNAKTLPHSKNSLTE 530
MKTVCVLTLGWLLFDSE+TFKNI GM +A+VGMVIYSWAV++EKQ NAK+ PH K+S+TE
Sbjct: 263 MKTVCVLTLGWLLFDSEMTFKNIAGMAIAIVGMVIYSWAVDIEKQRNAKSTPHGKHSMTE 322
Query: 529 EEIRLLKAGVENNPLKDVELGEAK 458
+EI+LLK GVE+ LKDVELG+ K
Sbjct: 323 DEIKLLKEGVEHIDLKDVELGDTK 346
>dbj|BAC42299.1| unknown protein [Arabidopsis thaliana]
Length = 312
Score = 330 bits (846), Expect = 2e-89
Identities = 158/204 (77%), Positives = 182/204 (88%)
Frame = -3
Query: 1069 GVGVCTVTDVKVNLKGFVCACLAILSTSLQQISIGSLQKKYSIGSFELLSKTAPIQAVSL 890
GVG+CTVTDVKVN KGF+CAC A+ STSLQQISIGSLQKKYSIGSFELLSKTAPIQA+SL
Sbjct: 108 GVGICTVTDVKVNAKGFICACTAVFSTSLQQISIGSLQKKYSIGSFELLSKTAPIQAISL 167
Query: 889 LVLGPFIDYYLSAKLITNYKMSSGAILFILLSCTLAVFCNVSQYLCIGRFSAVSFQVLGH 710
L+ GPF+DY+LS + I+ YKM+ A+L ILLSC LAVFCN+SQYLCIGRFSA SFQVLGH
Sbjct: 168 LIFGPFVDYFLSGRFISTYKMTYSAMLCILLSCALAVFCNISQYLCIGRFSATSFQVLGH 227
Query: 709 MKTVCVLTLGWLLFDSELTFKNIMGMVLAVVGMVIYSWAVEVEKQSNAKTLPHSKNSLTE 530
MKTVCVLTLGWL+FDSE+TFKNI GMVLAVVGMVIYSWAVE+EKQ +K +PH K+S+TE
Sbjct: 228 MKTVCVLTLGWLIFDSEMTFKNIAGMVLAVVGMVIYSWAVELEKQRKSKVIPHGKHSMTE 287
Query: 529 EEIRLLKAGVENNPLKDVELGEAK 458
+EI+LLK G+E+ LKD+ELG K
Sbjct: 288 DEIKLLKEGIEHMDLKDMELGNNK 311
>ref|NP_564133.1| expressed protein; protein id: At1g21070.1, supported by cDNA:
108746. [Arabidopsis thaliana] gi|25354485|pir||G86343
hypothetical protein T22I11.10 - Arabidopsis thaliana
gi|8886994|gb|AAF80654.1|AC012190_10 Strong similarity to
a hypothetical protein F28O16.4 gi|6143887 from
Arabidopsis thaliana gb|AC010718. It contains a integral
membrane protein domain PF|00892
Length = 348
Score = 330 bits (846), Expect = 2e-89
Identities = 158/204 (77%), Positives = 182/204 (88%)
Frame = -3
Query: 1069 GVGVCTVTDVKVNLKGFVCACLAILSTSLQQISIGSLQKKYSIGSFELLSKTAPIQAVSL 890
GVG+CTVTDVKVN KGF+CAC A+ STSLQQISIGSLQKKYSIGSFELLSKTAPIQA+SL
Sbjct: 144 GVGICTVTDVKVNAKGFICACTAVFSTSLQQISIGSLQKKYSIGSFELLSKTAPIQAISL 203
Query: 889 LVLGPFIDYYLSAKLITNYKMSSGAILFILLSCTLAVFCNVSQYLCIGRFSAVSFQVLGH 710
L+ GPF+DY+LS + I+ YKM+ A+L ILLSC LAVFCN+SQYLCIGRFSA SFQVLGH
Sbjct: 204 LIFGPFVDYFLSGRFISTYKMTYSAMLCILLSCALAVFCNISQYLCIGRFSATSFQVLGH 263
Query: 709 MKTVCVLTLGWLLFDSELTFKNIMGMVLAVVGMVIYSWAVEVEKQSNAKTLPHSKNSLTE 530
MKTVCVLTLGWL+FDSE+TFKNI GMVLAVVGMVIYSWAVE+EKQ +K +PH K+S+TE
Sbjct: 264 MKTVCVLTLGWLIFDSEMTFKNIAGMVLAVVGMVIYSWAVELEKQRKSKVIPHGKHSMTE 323
Query: 529 EEIRLLKAGVENNPLKDVELGEAK 458
+EI+LLK G+E+ LKD+ELG K
Sbjct: 324 DEIKLLKEGIEHMDLKDMELGNNK 347
>gb|AAM61035.1| unknown [Arabidopsis thaliana]
Length = 348
Score = 328 bits (841), Expect = 9e-89
Identities = 157/204 (76%), Positives = 181/204 (87%)
Frame = -3
Query: 1069 GVGVCTVTDVKVNLKGFVCACLAILSTSLQQISIGSLQKKYSIGSFELLSKTAPIQAVSL 890
GVG+CTVTDVK N KGF+CAC A+ STSLQQISIGSLQKKYSIGSFELLSKTAPIQA+SL
Sbjct: 144 GVGICTVTDVKFNAKGFICACTAVFSTSLQQISIGSLQKKYSIGSFELLSKTAPIQAISL 203
Query: 889 LVLGPFIDYYLSAKLITNYKMSSGAILFILLSCTLAVFCNVSQYLCIGRFSAVSFQVLGH 710
L+ GPF+DY+LS + I+ YKM+ A+L ILLSC LAVFCN+SQYLCIGRFSA SFQVLGH
Sbjct: 204 LIFGPFVDYFLSGRFISTYKMTYSAMLCILLSCALAVFCNISQYLCIGRFSATSFQVLGH 263
Query: 709 MKTVCVLTLGWLLFDSELTFKNIMGMVLAVVGMVIYSWAVEVEKQSNAKTLPHSKNSLTE 530
MKTVCVLTLGWL+FDSE+TFKNI GMVLAVVGMVIYSWAVE+EKQ +K +PH K+S+TE
Sbjct: 264 MKTVCVLTLGWLIFDSEMTFKNIAGMVLAVVGMVIYSWAVELEKQRKSKVIPHGKHSMTE 323
Query: 529 EEIRLLKAGVENNPLKDVELGEAK 458
+EI+LLK G+E+ LKD+ELG K
Sbjct: 324 DEIKLLKEGIEHMDLKDMELGNNK 347
>ref|NP_199057.1| putative protein; protein id: At5g42420.1 [Arabidopsis thaliana]
gi|9759478|dbj|BAB10483.1|
gb|AAF04433.1~gene_id:MDH9.11~strong similarity to
unknown protein [Arabidopsis thaliana]
Length = 350
Score = 313 bits (802), Expect = 3e-84
Identities = 157/206 (76%), Positives = 174/206 (84%), Gaps = 2/206 (0%)
Frame = -3
Query: 1069 GVGVCTVTDVKVNLKGFVCACLAILSTSLQQISIGSLQKKYSIGSFELLSKTAPIQAVSL 890
GVG+CTVTDVKVN KGF+CAC+AI S+SLQQI IGSLQKKYSIGSFELLSKTAPIQA SL
Sbjct: 144 GVGICTVTDVKVNAKGFICACVAIFSSSLQQILIGSLQKKYSIGSFELLSKTAPIQAFSL 203
Query: 889 LVLGPFIDYYLSAKLITNYKMSSGAILFILLSCTLAVFCNVSQYLCIGRFSAVSFQVLGH 710
LV+GP +DY LS K I Y MSSG LFILLSC LAVFCN+SQYLCIGRFSAVSFQV+GH
Sbjct: 204 LVVGPLVDYLLSGKFIMKYNMSSGCFLFILLSCGLAVFCNISQYLCIGRFSAVSFQVIGH 263
Query: 709 MKTVCVLTLGWLLFDSELTFKNIMGMVLAVVGMVIYSWAVEVEKQS--NAKTLPHSKNSL 536
MKTVC+LTLGWLLFDS +TFKN+ GM++A+VGMVIYSWA+E+EKQS AK L K+SL
Sbjct: 264 MKTVCILTLGWLLFDSAMTFKNVAGMIVAIVGMVIYSWAMELEKQSIIAAKALNSVKHSL 323
Query: 535 TEEEIRLLKAGVENNPLKDVELGEAK 458
TEEE LLK GVE KDVELG K
Sbjct: 324 TEEEFELLKEGVETTQSKDVELGRTK 349
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 886,916,024
Number of Sequences: 1393205
Number of extensions: 19181071
Number of successful extensions: 46493
Number of sequences better than 10.0: 114
Number of HSP's better than 10.0 without gapping: 44214
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 46403
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 64016183864
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)