Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003516A_C01 KMC003516A_c01
(617 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
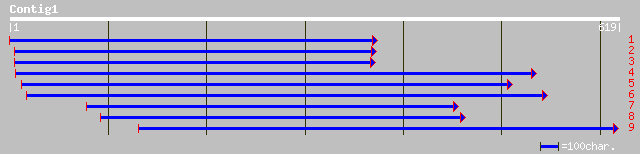
ref|NP_680213.1| Expressed protein; protein id: At5g23575.1, sup... 139 3e-32
dbj|BAA97243.1| gb|AAC97420.1~gene_id:MQM1.16~strong similarity ... 139 3e-32
ref|NP_196467.1| cleft lip and palate associated transmembrane p... 137 1e-31
ref|NP_608819.1| CG3702-PA [Drosophila melanogaster] gi|15292391... 110 1e-23
ref|NP_509196.1| Putative endoplasmic reticulum membrane protein... 107 2e-22
>ref|NP_680213.1| Expressed protein; protein id: At5g23575.1, supported by cDNA:
gi_17381231 [Arabidopsis thaliana]
gi|17381232|gb|AAL36035.1| AT5g08500/MAH20_6
[Arabidopsis thaliana] gi|23506007|gb|AAN28863.1|
At5g08500/MAH20_6 [Arabidopsis thaliana]
Length = 593
Score = 139 bits (350), Expect = 3e-32
Identities = 67/84 (79%), Positives = 73/84 (86%)
Frame = -1
Query: 617 WRQMTYKFLNTIIDDLFAFVIKMPTLHRLSVFRDDLIFLIYVYQRWIYPVDKKRVNEFGF 438
WRQMTYKFLNTIIDDLFAFVIKMP LHRLSVFRDD+IFLIY+YQRW+YPVDK RVNEFGF
Sbjct: 511 WRQMTYKFLNTIIDDLFAFVIKMPILHRLSVFRDDVIFLIYLYQRWVYPVDKTRVNEFGF 570
Query: 437 GGEDDQAVVSAETDAATEEEKKTN 366
GGED+ A T+ EE+KKTN
Sbjct: 571 GGEDETAEKKLITE-KEEEDKKTN 593
>dbj|BAA97243.1| gb|AAC97420.1~gene_id:MQM1.16~strong similarity to unknown protein
[Arabidopsis thaliana]
Length = 583
Score = 139 bits (350), Expect = 3e-32
Identities = 67/84 (79%), Positives = 73/84 (86%)
Frame = -1
Query: 617 WRQMTYKFLNTIIDDLFAFVIKMPTLHRLSVFRDDLIFLIYVYQRWIYPVDKKRVNEFGF 438
WRQMTYKFLNTIIDDLFAFVIKMP LHRLSVFRDD+IFLIY+YQRW+YPVDK RVNEFGF
Sbjct: 501 WRQMTYKFLNTIIDDLFAFVIKMPILHRLSVFRDDVIFLIYLYQRWVYPVDKTRVNEFGF 560
Query: 437 GGEDDQAVVSAETDAATEEEKKTN 366
GGED+ A T+ EE+KKTN
Sbjct: 561 GGEDETAEKKLITE-KEEEDKKTN 583
>ref|NP_196467.1| cleft lip and palate associated transmembrane protein-like; protein
id: At5g08500.1, supported by cDNA: gi_20466779
[Arabidopsis thaliana] gi|9759344|dbj|BAB09999.1| cleft
lip and palate associated transmembrane protein-like
[Arabidopsis thaliana] gi|20466780|gb|AAM20707.1| cleft
lip and palate associated transmembrane protein-like
[Arabidopsis thaliana]
Length = 590
Score = 137 bits (345), Expect = 1e-31
Identities = 65/84 (77%), Positives = 73/84 (86%)
Frame = -1
Query: 617 WRQMTYKFLNTIIDDLFAFVIKMPTLHRLSVFRDDLIFLIYVYQRWIYPVDKKRVNEFGF 438
WRQMTYKFLNTIIDDLFAFVIKMP LHRLSVFRDD+IFLIY+YQRW+YPVDK RVNEFGF
Sbjct: 509 WRQMTYKFLNTIIDDLFAFVIKMPMLHRLSVFRDDVIFLIYLYQRWVYPVDKTRVNEFGF 568
Query: 437 GGEDDQAVVSAETDAATEEEKKTN 366
GGED+ V + + E++KKTN
Sbjct: 569 GGEDES--VDTKKISDQEDDKKTN 590
>ref|NP_608819.1| CG3702-PA [Drosophila melanogaster] gi|15292391|gb|AAK93464.1|
LP05054p [Drosophila melanogaster]
gi|22953827|gb|AAF51035.2| CG3702-PA [Drosophila
melanogaster]
Length = 641
Score = 110 bits (275), Expect = 1e-23
Identities = 49/77 (63%), Positives = 60/77 (77%)
Frame = -1
Query: 617 WRQMTYKFLNTIIDDLFAFVIKMPTLHRLSVFRDDLIFLIYVYQRWIYPVDKKRVNEFGF 438
WR MTYKFLNT IDD+FAFVIKMPT++RL FRDD+IF +++YQRW Y VD KRVNEFGF
Sbjct: 541 WRMMTYKFLNTFIDDIFAFVIKMPTMYRLGCFRDDIIFFVFLYQRWQYRVDLKRVNEFGF 600
Query: 437 GGEDDQAVVSAETDAAT 387
GE +Q + + + T
Sbjct: 601 SGEMEQQHAAQKLEGKT 617
>ref|NP_509196.1| Putative endoplasmic reticulum membrane protein family member, with
at least 5 transmembrane domains, of eukaryotic origin
[Caenorhabditis elegans]
gi|14573912|gb|AAK68233.1|AC006615_7 Hypothetical
protein C36B7.6 [Caenorhabditis elegans]
Length = 670
Score = 107 bits (266), Expect = 2e-22
Identities = 51/89 (57%), Positives = 63/89 (70%), Gaps = 7/89 (7%)
Frame = -1
Query: 617 WRQMTYKFLNTIIDDLFAFVIKMPTLHRLSVFRDDLIFLIYVYQRWIYPVDKKRVNEFGF 438
WR +TYKF+NT IDDLFAFVIKMP L+R+ FRDD+IFLIY+YQRWIY VD R+NE+G
Sbjct: 579 WRMLTYKFINTFIDDLFAFVIKMPLLYRIGCFRDDIIFLIYLYQRWIYRVDPTRMNEYGT 638
Query: 437 GGEDDQAV-------VSAETDAATEEEKK 372
GE + V +A++D EE K
Sbjct: 639 SGEHEHGVPEPVEGTETAKSDTPAIEESK 667
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 486,067,274
Number of Sequences: 1393205
Number of extensions: 9867436
Number of successful extensions: 24108
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 23201
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24085
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 24733321959
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)