Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003508A_C01 KMC003508A_c01
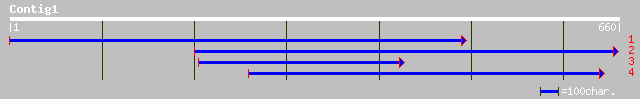
(659 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|EAA22565.1| Var1p [Plasmodium yoelii yoelii] 34 2.0
gb|EAA42616.1| GLP_487_61862_64360 [Giardia lamblia ATCC 50803] 33 2.6
ref|NP_195294.1| putative protein; protein id: At4g35690.1 [Arab... 33 2.6
ref|XP_225693.1| similar to zinc finger protein 236 [Homo sapien... 33 4.4
ref|NP_048091.1| ORF MSV020 hypothetical protein [Melanoplus san... 30 9.2
>gb|EAA22565.1| Var1p [Plasmodium yoelii yoelii]
Length = 548
Score = 33.9 bits (76), Expect = 2.0
Identities = 21/79 (26%), Positives = 37/79 (46%)
Frame = +3
Query: 123 FFFFFIK*ILFTLLENYTIVQHY*SNYKLKKLYINNQTMDFWGLYLESKDVNQFRSLSHY 302
F FFF+K F E Y + +Y +N K+ Y NN + K+++ SL +
Sbjct: 430 FLFFFLKYTQFVQFEKYNSIYNYTTNDKILVNYDNNDIYSNLNEERKKKNIDCDPSLIPH 489
Query: 303 ISKQIIHLLAHAIKKLLKF 359
+S I + ++ + L K+
Sbjct: 490 VSHNEISIYSNILSILKKY 508
>gb|EAA42616.1| GLP_487_61862_64360 [Giardia lamblia ATCC 50803]
Length = 832
Score = 33.5 bits (75), Expect = 2.6
Identities = 23/56 (41%), Positives = 31/56 (55%)
Frame = -1
Query: 431 LLKLLLLMK*KDMKKPLNLQTKLHEFEKLLDGMSKEVDNLFADVMTQRSELINVFR 264
L + LLM+ KD + + L LH EK LD + KEV N F++V+ EL N R
Sbjct: 568 LTDIKLLMEEKD-RNLIQLTEDLHRKEKELDALVKEVKNAFSEVL----ELTNTVR 618
>ref|NP_195294.1| putative protein; protein id: At4g35690.1 [Arabidopsis thaliana]
gi|7486647|pir||T04674 hypothetical protein F8D20.200 -
Arabidopsis thaliana gi|3367587|emb|CAA20039.1| putative
protein [Arabidopsis thaliana]
gi|7270520|emb|CAB80285.1| putative protein [Arabidopsis
thaliana]
Length = 284
Score = 33.5 bits (75), Expect = 2.6
Identities = 14/37 (37%), Positives = 24/37 (64%)
Frame = -1
Query: 380 NLQTKLHEFEKLLDGMSKEVDNLFADVMTQRSELINV 270
+LQ KL E E +DG K+++ LF ++ R+ L+N+
Sbjct: 245 DLQKKLEEVEMSIDGFEKKLEGLFRRLIRTRASLLNI 281
>ref|XP_225693.1| similar to zinc finger protein 236 [Homo sapiens] [Rattus norvegicus]
Length = 1007
Score = 32.7 bits (73), Expect = 4.4
Identities = 17/76 (22%), Positives = 35/76 (45%)
Frame = +2
Query: 287 ISESLHQQTDYPPPCSCHQEASQIHAALSASSVVSSCLFTSSEAATSATKLFSSWISPNI 466
++++ QQT +PP ++ ALS S + L S A + + W+ P
Sbjct: 919 VTDTYSQQTSFPPVQQLQDSSTLESQALSTSFHQQNLLQVPSSDAINVVSVEKGWVVPQS 978
Query: 467 GIFSINCSLILKSRVA 514
++C+ I+++ +A
Sbjct: 979 ADICLSCAGIIQNALA 994
>ref|NP_048091.1| ORF MSV020 hypothetical protein [Melanoplus sanguinipes
entomopoxvirus] gi|11289298|pir||T28181 hypothetical
protein ORF20 - Melanoplus sanguinipes entomopoxvirus
(isolate Tuscon) gi|4049653|gb|AAC97613.1| ORF MSV020
hypothetical protein [Melanoplus sanguinipes
entomopoxvirus]
Length = 175
Score = 29.6 bits (65), Expect(2) = 9.2
Identities = 20/66 (30%), Positives = 36/66 (54%)
Frame = +3
Query: 162 LENYTIVQHY*SNYKLKKLYINNQTMDFWGLYLESKDVNQFRSLSHYISKQIIHLLAHAI 341
+ENY V H + + L K IN + ++ + + E+ +QF+ L +++ K I + H I
Sbjct: 1 MENYN-VSHITNMFNLLKTPINYKRLEKYFIKNENFSKHQFKELFNFMQKYIYKI--HNI 57
Query: 342 KKLLKF 359
K+L F
Sbjct: 58 KELYLF 63
Score = 20.8 bits (42), Expect(2) = 9.2
Identities = 7/19 (36%), Positives = 12/19 (62%)
Frame = +1
Query: 394 MSFYFIRSSNFSNKTVQFL 450
+ +YFI++ NFS + L
Sbjct: 98 LEYYFIKNENFSKYQFKLL 116
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 534,656,964
Number of Sequences: 1393205
Number of extensions: 11005296
Number of successful extensions: 37505
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 34618
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37431
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28289785200
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)