Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
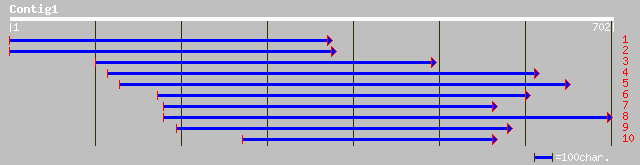
Query= KMC003488A_C01 KMC003488A_c01
(701 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||C85342 hypothetical protein AT4g29330 [imported] - Arabidop... 116 3e-25
ref|NP_194662.2| putative protein; protein id: At4g29330.1, supp... 116 3e-25
emb|CAB97005.1| putative NADH oxidoreductase [Zea mays] 107 1e-22
gb|AAO20072.1| putative Der1-like protein [Oryza sativa (japonic... 47 3e-04
ref|NP_192395.1| unknown protein; protein id: At4g04860.1, suppo... 46 4e-04
>pir||C85342 hypothetical protein AT4g29330 [imported] - Arabidopsis thaliana
gi|7269831|emb|CAB79691.1| putative protein [Arabidopsis
thaliana]
Length = 281
Score = 116 bits (291), Expect = 3e-25
Identities = 63/105 (60%), Positives = 68/105 (64%), Gaps = 20/105 (19%)
Frame = -1
Query: 701 DVIFGSPIIPDLLGIIAGHLYYFLTVLHPLAGGKNILKTPMWVHKLVARWRIGAPAVVSR 522
DVIFGSPI+PDLLGIIAGHLYYFLTVLHPLA GKN LKTP WV+K+VARWRIGAP R
Sbjct: 177 DVIFGSPIMPDLLGIIAGHLYYFLTVLHPLATGKNYLKTPKWVNKIVARWRIGAPVASVR 236
Query: 521 --------------------AQPVNNARQESSSGVFRGRSYRLNE 447
A A ESS+ FRGRSYRL +
Sbjct: 237 QAGGVGAAGPGAGGGVGGGGAYSSARAPPESSNTAFRGRSYRLTD 281
>ref|NP_194662.2| putative protein; protein id: At4g29330.1, supported by cDNA:
gi_17380685, supported by cDNA: gi_20465888 [Arabidopsis
thaliana] gi|17380686|gb|AAL36173.1| unknown protein
[Arabidopsis thaliana] gi|20465889|gb|AAM20097.1|
unknown protein [Arabidopsis thaliana]
Length = 266
Score = 116 bits (291), Expect = 3e-25
Identities = 63/105 (60%), Positives = 68/105 (64%), Gaps = 20/105 (19%)
Frame = -1
Query: 701 DVIFGSPIIPDLLGIIAGHLYYFLTVLHPLAGGKNILKTPMWVHKLVARWRIGAPAVVSR 522
DVIFGSPI+PDLLGIIAGHLYYFLTVLHPLA GKN LKTP WV+K+VARWRIGAP R
Sbjct: 162 DVIFGSPIMPDLLGIIAGHLYYFLTVLHPLATGKNYLKTPKWVNKIVARWRIGAPVASVR 221
Query: 521 --------------------AQPVNNARQESSSGVFRGRSYRLNE 447
A A ESS+ FRGRSYRL +
Sbjct: 222 QAGGVGAAGPGAGGGVGGGGAYSSARAPPESSNTAFRGRSYRLTD 266
>emb|CAB97005.1| putative NADH oxidoreductase [Zea mays]
Length = 259
Score = 107 bits (268), Expect = 1e-22
Identities = 54/85 (63%), Positives = 62/85 (72%)
Frame = -1
Query: 701 DVIFGSPIIPDLLGIIAGHLYYFLTVLHPLAGGKNILKTPMWVHKLVARWRIGAPAVVSR 522
DVIFGSP++P LLGI+ GHLYY+ VL PLA GK+ LKTP WVHK+VAR+RIG A
Sbjct: 178 DVIFGSPLMPGLLGIMVGHLYYYFAVLDPLATGKSYLKTPKWVHKIVARFRIGMQANSPV 237
Query: 521 AQPVNNARQESSSGVFRGRSYRLNE 447
P N S SGVFRGRSYRLN+
Sbjct: 238 RPPANG---NSGSGVFRGRSYRLNQ 259
>gb|AAO20072.1| putative Der1-like protein [Oryza sativa (japonica cultivar-group)]
Length = 249
Score = 47.0 bits (110), Expect = 3e-04
Identities = 22/47 (46%), Positives = 31/47 (65%)
Frame = -1
Query: 698 VIFGSPIIPDLLGIIAGHLYYFLTVLHPLAGGKNILKTPMWVHKLVA 558
++ GS DLLG+IAGH+YYFL ++P G+ LKTP ++ L A
Sbjct: 173 ILVGSSTWVDLLGMIAGHVYYFLEDVYPRMTGRRPLKTPSFIKALFA 219
>ref|NP_192395.1| unknown protein; protein id: At4g04860.1, supported by cDNA:
gi_14488072, supported by cDNA: gi_20147126 [Arabidopsis
thaliana] gi|25407297|pir||B85061 hypothetical protein
AT4g04860 [imported] - Arabidopsis thaliana
gi|4115936|gb|AAD03446.1| T4B21.2 gene product
[Arabidopsis thaliana] gi|7267244|emb|CAB80851.1|
predicted protein of unknown function [Arabidopsis
thaliana] gi|14488073|gb|AAK63857.1|AF389284_1
AT4g04860/T4B21_2 [Arabidopsis thaliana]
gi|20147127|gb|AAM10280.1| AT4g04860/T4B21_2
[Arabidopsis thaliana]
Length = 244
Score = 46.2 bits (108), Expect = 4e-04
Identities = 26/66 (39%), Positives = 37/66 (55%)
Frame = -1
Query: 698 VIFGSPIIPDLLGIIAGHLYYFLTVLHPLAGGKNILKTPMWVHKLVARWRIGAPAVVSRA 519
++ G+ DLLG+IAGH YYFL ++P + LKTP ++ L A P VV+R
Sbjct: 173 ILVGASAWVDLLGMIAGHAYYFLAEVYPRMTNRRPLKTPSFLKALFA----DEPVVVARP 228
Query: 518 QPVNNA 501
+ V A
Sbjct: 229 EDVRFA 234
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 615,570,563
Number of Sequences: 1393205
Number of extensions: 13780393
Number of successful extensions: 33435
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 32489
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33418
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 32250355128
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)