Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003472A_C01 KMC003472A_c01
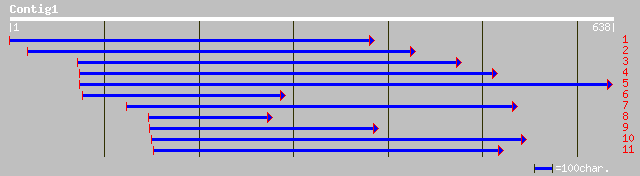
(632 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK98698.1|AC069158_10 Putative GATA-1 zinc finger protein [O... 94 5e-25
ref|NP_191612.1| GATA zinc finger protein; protein id: At3g60530... 92 1e-24
gb|AAK55449.1|AC069300_4 putative transcription factor [Oryza sa... 91 4e-24
ref|NP_182031.1| GATA zinc finger protein; protein id: At2g45050... 91 4e-24
ref|NP_195015.1| GATA zinc finger protein; protein id: At4g32890... 92 7e-24
>gb|AAK98698.1|AC069158_10 Putative GATA-1 zinc finger protein [Oryza sativa]
Length = 418
Score = 94.4 bits (233), Expect(2) = 5e-25
Identities = 42/49 (85%), Positives = 45/49 (91%)
Frame = -1
Query: 521 GPLGPKTLCNACGVRYKSGRLHPEYRPAKSPTFVSFLHSNSHKKVMEMR 375
GPLGPKTLCNACGVRYKSGRL PEYRPA SPTF+ +HSNSHKKV+EMR
Sbjct: 349 GPLGPKTLCNACGVRYKSGRLFPEYRPAASPTFMPSIHSNSHKKVVEMR 397
Score = 42.0 bits (97), Expect(2) = 5e-25
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 4/39 (10%)
Frame = -3
Query: 609 DGEVSNNGQNPMP----RRCTHCLSQRTPQWRAGAIGAK 505
D + G +P RRCTHC ++TPQWRAG +G K
Sbjct: 316 DADYEEGGALALPPGTVRRCTHCQIEKTPQWRAGPLGPK 354
>ref|NP_191612.1| GATA zinc finger protein; protein id: At3g60530.1, supported by
cDNA: gi_14190406, supported by cDNA: gi_14517394,
supported by cDNA: gi_15215890 [Arabidopsis thaliana]
gi|11282352|pir||T47864 GATA transcription factor 4 -
Arabidopsis thaliana gi|2959736|emb|CAA74002.1|
homologous to GATA-binding transcription factors
[Arabidopsis thaliana] gi|7288001|emb|CAB81839.1| GATA
transcription factor 4 [Arabidopsis thaliana]
gi|14190407|gb|AAK55684.1|AF378881_1 AT3g60530/T8B10_190
[Arabidopsis thaliana] gi|14517395|gb|AAK62588.1|
AT3g60530/T8B10_190 [Arabidopsis thaliana]
gi|15215891|gb|AAK91489.1| AT3g60530/T8B10_190
[Arabidopsis thaliana]
Length = 240
Score = 92.4 bits (228), Expect(2) = 1e-24
Identities = 42/49 (85%), Positives = 44/49 (89%)
Frame = -1
Query: 521 GPLGPKTLCNACGVRYKSGRLHPEYRPAKSPTFVSFLHSNSHKKVMEMR 375
GPLGPKTLCNACGVRYKSGRL PEYRPA SPTFV HSNSH+KVME+R
Sbjct: 174 GPLGPKTLCNACGVRYKSGRLVPEYRPASSPTFVLTQHSNSHRKVMELR 222
Score = 42.4 bits (98), Expect(2) = 1e-24
Identities = 15/22 (68%), Positives = 18/22 (81%)
Frame = -3
Query: 570 RRCTHCLSQRTPQWRAGAIGAK 505
RRCTHC S++TPQWR G +G K
Sbjct: 158 RRCTHCASEKTPQWRTGPLGPK 179
>gb|AAK55449.1|AC069300_4 putative transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 387
Score = 90.9 bits (224), Expect(2) = 4e-24
Identities = 41/49 (83%), Positives = 44/49 (89%)
Frame = -1
Query: 521 GPLGPKTLCNACGVRYKSGRLHPEYRPAKSPTFVSFLHSNSHKKVMEMR 375
GPLGPKTLCNACGVR+KSGRL PEYRPA SPTFV HSNSH+KVME+R
Sbjct: 292 GPLGPKTLCNACGVRFKSGRLMPEYRPAASPTFVLTQHSNSHRKVMELR 340
Score = 42.4 bits (98), Expect(2) = 4e-24
Identities = 15/22 (68%), Positives = 18/22 (81%)
Frame = -3
Query: 570 RRCTHCLSQRTPQWRAGAIGAK 505
RRCTHC S++TPQWR G +G K
Sbjct: 276 RRCTHCASEKTPQWRTGPLGPK 297
>ref|NP_182031.1| GATA zinc finger protein; protein id: At2g45050.1 [Arabidopsis
thaliana] gi|25352345|pir||T52104 GATA-binding
transcription factor homolog 2 [imported] - Arabidopsis
thaliana gi|2959732|emb|CAA74000.1| homologous to
GATA-binding transcription factors [Arabidopsis
thaliana] gi|24030302|gb|AAN41321.1| putative GATA-type
zinc finger transcription factor [Arabidopsis thaliana]
Length = 264
Score = 90.9 bits (224), Expect(2) = 4e-24
Identities = 41/49 (83%), Positives = 44/49 (89%)
Frame = -1
Query: 521 GPLGPKTLCNACGVRYKSGRLHPEYRPAKSPTFVSFLHSNSHKKVMEMR 375
GPLGPKTLCNACGVR+KSGRL PEYRPA SPTFV HSNSH+KVME+R
Sbjct: 195 GPLGPKTLCNACGVRFKSGRLVPEYRPASSPTFVLTQHSNSHRKVMELR 243
Score = 42.4 bits (98), Expect(2) = 4e-24
Identities = 15/22 (68%), Positives = 18/22 (81%)
Frame = -3
Query: 570 RRCTHCLSQRTPQWRAGAIGAK 505
RRCTHC S++TPQWR G +G K
Sbjct: 179 RRCTHCASEKTPQWRTGPLGPK 200
>ref|NP_195015.1| GATA zinc finger protein; protein id: At4g32890.1 [Arabidopsis
thaliana] gi|7486206|pir||T05297 hypothetical protein
F26P21.10 - Arabidopsis thaliana
gi|3688170|emb|CAA21198.1| putative protein [Arabidopsis
thaliana] gi|7270236|emb|CAB80006.1| putative protein
[Arabidopsis thaliana] gi|26449440|dbj|BAC41847.1|
unknown protein [Arabidopsis thaliana]
Length = 308
Score = 92.0 bits (227), Expect(2) = 7e-24
Identities = 41/49 (83%), Positives = 44/49 (89%)
Frame = -1
Query: 521 GPLGPKTLCNACGVRYKSGRLHPEYRPAKSPTFVSFLHSNSHKKVMEMR 375
GP+GPKTLCNACGVRYKSGRL PEYRPA SPTFV HSNSH+KVME+R
Sbjct: 213 GPMGPKTLCNACGVRYKSGRLVPEYRPASSPTFVMARHSNSHRKVMELR 261
Score = 40.4 bits (93), Expect(2) = 7e-24
Identities = 18/42 (42%), Positives = 28/42 (65%), Gaps = 1/42 (2%)
Frame = -3
Query: 627 VKEEFEDGEVSNN-GQNPMPRRCTHCLSQRTPQWRAGAIGAK 505
VKE+ G++ + G++ RRC HC +++TPQWR G +G K
Sbjct: 177 VKEQDFAGDMDVDCGESGGGRRCLHCATEKTPQWRTGPMGPK 218
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 575,041,828
Number of Sequences: 1393205
Number of extensions: 12751898
Number of successful extensions: 34851
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 33434
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34823
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26154777244
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)