Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003469A_C01 KMC003469A_c01
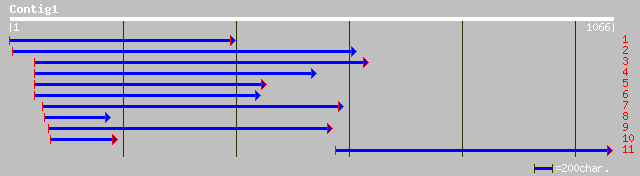
(1066 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC43324.1| unknown protein [Arabidopsis thaliana] 299 6e-80
pir||G71431 hypothetical protein - Arabidopsis thaliana gi|22450... 256 6e-67
dbj|BAC22265.1| arm repeat containing protein homolog-like [Oryz... 251 1e-65
ref|NP_566136.1| expressed protein; protein id: At3g01400.1, sup... 196 5e-49
gb|AAL16172.1|AF428404_1 AT3g01400/T13O15_4 [Arabidopsis thalian... 196 7e-49
>dbj|BAC43324.1| unknown protein [Arabidopsis thaliana]
Length = 472
Score = 299 bits (765), Expect = 6e-80
Identities = 159/191 (83%), Positives = 173/191 (90%), Gaps = 2/191 (1%)
Frame = -2
Query: 1065 ETSKQNAACALLSLALVEENRSSIGASGAIPPLVSLLINGSSRGKKDALTTLYKLCSVKL 886
ETSKQNAACALLSLAL+EEN+ SIGA GAIPPLVSLL+NGS RGKKDALT LYKLC+++
Sbjct: 280 ETSKQNAACALLSLALLEENKGSIGACGAIPPLVSLLLNGSCRGKKDALTALYKLCTLQQ 339
Query: 885 NKERAVNAGVVKPLVDLVAEQGTGLAEKAMVVLNSLAAVQDGKDAIVEEGGIAALVEAIE 706
NKERAV AG VKPLVDLVAE+GTG+AEKAMVVL+SLAA+ DGK+AIVEEGGIAALVEAIE
Sbjct: 340 NKERAVTAGAVKPLVDLVAEEGTGMAEKAMVVLSSLAAIDDGKEAIVEEGGIAALVEAIE 399
Query: 705 DGSVKGKEFAVLTLLQLCVDSVRNRGLLVREGGIPPLVSLSQSG--TPRAKHKAETLLRY 532
DGSVKGKEFA+LTLLQLC DSVRNRGLLVREG IPPLV LSQSG + RAK KAE LL Y
Sbjct: 400 DGSVKGKEFAILTLLQLCSDSVRNRGLLVREGAIPPLVGLSQSGSVSVRAKRKAERLLGY 459
Query: 531 LRESRQEASSS 499
LRE R+EASSS
Sbjct: 460 LREPRKEASSS 470
Score = 66.2 bits (160), Expect = 8e-10
Identities = 60/178 (33%), Positives = 88/178 (48%), Gaps = 2/178 (1%)
Frame = -2
Query: 1056 KQNAACALLSLALVE-ENRSSIGASGAIPPLVSLLINGSSRGKKDALTTLYKLCSVKLNK 880
K++AA L LA +NR IG SGAI L+ LL ++ A+T L L NK
Sbjct: 200 KRSAAAKLRLLAKNRADNRVLIGESGAIQALIPLLRCNDPWTQERAVTALLNLSLHDQNK 259
Query: 879 ERAVNAGVVKPLVDLVAEQGTGLAEKAMVVLNSLAAVQDGKDAIVEEGGIAALVEAIEDG 700
G +K LV ++ + A L SLA +++ K +I G I LV + +G
Sbjct: 260 AVIAAGGAIKSLVWVLKTGTETSKQNAACALLSLALLEENKGSIGACGAIPPLVSLLLNG 319
Query: 699 SVKGKEFAVLTLLQLCVDSVRNRGLLVREGGIPPLVSL-SQSGTPRAKHKAETLLRYL 529
S +GK+ A+ L +LC +N+ V G + PLV L ++ GT A+ KA +L L
Sbjct: 320 SCRGKKDALTALYKLCT-LQQNKERAVTAGAVKPLVDLVAEEGTGMAE-KAMVVLSSL 375
>pir||G71431 hypothetical protein - Arabidopsis thaliana
gi|2245005|emb|CAB10425.1| hypothetical protein
[Arabidopsis thaliana] gi|7268399|emb|CAB78691.1|
hypothetical protein [Arabidopsis thaliana]
Length = 459
Score = 256 bits (653), Expect = 6e-67
Identities = 132/165 (80%), Positives = 148/165 (89%)
Frame = -2
Query: 1032 LSLALVEENRSSIGASGAIPPLVSLLINGSSRGKKDALTTLYKLCSVKLNKERAVNAGVV 853
L LAL+EEN+ SIGA GAIPPLVSLL+NGS RGKKDALTTLYKLC+++ NKERAV AG V
Sbjct: 160 LGLALLEENKGSIGACGAIPPLVSLLLNGSCRGKKDALTTLYKLCTLQQNKERAVTAGAV 219
Query: 852 KPLVDLVAEQGTGLAEKAMVVLNSLAAVQDGKDAIVEEGGIAALVEAIEDGSVKGKEFAV 673
KPLVDLVAE+GTG+AEKAMVVL+SLAA+ DGK+AIVEEGGIAALVEAIEDGSVKGKEFA+
Sbjct: 220 KPLVDLVAEEGTGMAEKAMVVLSSLAAIDDGKEAIVEEGGIAALVEAIEDGSVKGKEFAI 279
Query: 672 LTLLQLCVDSVRNRGLLVREGGIPPLVSLSQSGTPRAKHKAETLL 538
LTLLQLC DSVRNRGLLVREG IPPLV LSQSG+ + K + +L
Sbjct: 280 LTLLQLCSDSVRNRGLLVREGAIPPLVGLSQSGSVSVRAKRKNVL 324
>dbj|BAC22265.1| arm repeat containing protein homolog-like [Oryza sativa (japonica
cultivar-group)]
Length = 495
Score = 251 bits (642), Expect = 1e-65
Identities = 129/183 (70%), Positives = 154/183 (83%)
Frame = -2
Query: 1062 TSKQNAACALLSLALVEENRSSIGASGAIPPLVSLLINGSSRGKKDALTTLYKLCSVKLN 883
++KQNAACALLSL+ +EENR++IGA GAIPPLV+LL GS+RGKKDALTTLY+LCS + N
Sbjct: 268 SAKQNAACALLSLSGIEENRATIGACGAIPPLVALLSAGSTRGKKDALTTLYRLCSARRN 327
Query: 882 KERAVNAGVVKPLVDLVAEQGTGLAEKAMVVLNSLAAVQDGKDAIVEEGGIAALVEAIED 703
KERAV+AG V PL+ LV E+G+G +EKAMVVL SLA + +G+DA+VE GGI ALVE IED
Sbjct: 328 KERAVSAGAVVPLIHLVGERGSGTSEKAMVVLASLAGIVEGRDAVVEAGGIPALVETIED 387
Query: 702 GSVKGKEFAVLTLLQLCVDSVRNRGLLVREGGIPPLVSLSQSGTPRAKHKAETLLRYLRE 523
G + +EFAV+ LLQLC + RNR LLVREG IPPLV+LSQSG+ RAKHKAETLL YLRE
Sbjct: 388 GPAREREFAVVALLQLCSECPRNRALLVREGAIPPLVALSQSGSARAKHKAETLLGYLRE 447
Query: 522 SRQ 514
RQ
Sbjct: 448 QRQ 450
Score = 75.5 bits (184), Expect = 1e-12
Identities = 56/178 (31%), Positives = 85/178 (47%), Gaps = 1/178 (0%)
Frame = -2
Query: 1059 SKQNAACALLSLALVEEN-RSSIGASGAIPPLVSLLINGSSRGKKDALTTLYKLCSVKLN 883
+++ AA + LA + R IG SGAIP LV LL + ++ A+T L L + N
Sbjct: 186 ARRTAAARIRLLAKHRSDIRELIGVSGAIPALVPLLRSTDPVAQESAVTALLNLSLEERN 245
Query: 882 KERAVNAGVVKPLVDLVAEQGTGLAEKAMVVLNSLAAVQDGKDAIVEEGGIAALVEAIED 703
+ AG +KPLV + + A L SL+ +++ + I G I LV +
Sbjct: 246 RSAITAAGAIKPLVYALRTGTASAKQNAACALLSLSGIEENRATIGACGAIPPLVALLSA 305
Query: 702 GSVKGKEFAVLTLLQLCVDSVRNRGLLVREGGIPPLVSLSQSGTPRAKHKAETLLRYL 529
GS +GK+ A+ TL +LC + RN+ V G + PL+ L KA +L L
Sbjct: 306 GSTRGKKDALTTLYRLC-SARRNKERAVSAGAVVPLIHLVGERGSGTSEKAMVVLASL 362
Score = 40.8 bits (94), Expect = 0.036
Identities = 29/91 (31%), Positives = 42/91 (45%)
Frame = -2
Query: 765 DGKDAIVEEGGIAALVEAIEDGSVKGKEFAVLTLLQLCVDSVRNRGLLVREGGIPPLVSL 586
D ++ I G I ALV + +E AV LL L ++ RNR + G I PLV
Sbjct: 203 DIRELIGVSGAIPALVPLLRSTDPVAQESAVTALLNLSLEE-RNRSAITAAGAIKPLVYA 261
Query: 585 SQSGTPRAKHKAETLLRYLRESRQEASSSGS 493
++GT AK A L L + ++ G+
Sbjct: 262 LRTGTASAKQNAACALLSLSGIEENRATIGA 292
>ref|NP_566136.1| expressed protein; protein id: At3g01400.1, supported by cDNA:
34582., supported by cDNA: gi_16226453 [Arabidopsis
thaliana] gi|6692260|gb|AAF24610.1|AC010870_3 unknown
protein [Arabidopsis thaliana]
Length = 355
Score = 196 bits (498), Expect = 5e-49
Identities = 106/194 (54%), Positives = 141/194 (72%)
Frame = -2
Query: 1062 TSKQNAACALLSLALVEENRSSIGASGAIPPLVSLLINGSSRGKKDALTTLYKLCSVKLN 883
T+K+NAACALL L+ +EEN+ +IG SGAIP LV+LL G R KKDA T LY LCS K N
Sbjct: 161 TAKENAACALLRLSQIEENKVAIGRSGAIPLLVNLLETGGFRAKKDASTALYSLCSAKEN 220
Query: 882 KERAVNAGVVKPLVDLVAEQGTGLAEKAMVVLNSLAAVQDGKDAIVEEGGIAALVEAIED 703
K RAV +G++KPLV+L+A+ G+ + +K+ V++ L +V + K AIVEEGG+ LVE +E
Sbjct: 221 KIRAVQSGIMKPLVELMADFGSNMVDKSAFVMSLLMSVPESKPAIVEEGGVPVLVEIVEV 280
Query: 702 GSVKGKEFAVLTLLQLCVDSVRNRGLLVREGGIPPLVSLSQSGTPRAKHKAETLLRYLRE 523
G+ + KE AV LLQLC +SV R ++ REG IPPLV+LSQ+GT RAK KAE L+ LR+
Sbjct: 281 GTQRQKEMAVSILLQLCEESVVYRTMVAREGAIPPLVALSQAGTSRAKQKAEALIELLRQ 340
Query: 522 SRQEASSSGS*RSN 481
R + S+G RS+
Sbjct: 341 PR--SISNGGARSS 352
Score = 70.5 bits (171), Expect = 4e-11
Identities = 57/204 (27%), Positives = 87/204 (41%), Gaps = 41/204 (20%)
Frame = -2
Query: 1011 ENRSSIGASGAIPPLVSLLINGSSRGKKDALTTLYKLCSVKLNKERAVNAGVVKPLVDLV 832
ENR I +GAI PL+SL+ + + ++ +T + L NKE ++G +KPLV +
Sbjct: 96 ENRIKIAKAGAIKPLISLISSSDLQLQEYGVTAILNLSLCDENKESIASSGAIKPLVRAL 155
Query: 831 AEQGTGLA-EKAMVVLNSLAAVQDGKDAIVEEGGIAALVEAIEDGSVKGKEFAVLTLLQL 655
+ GT A E A L L+ +++ K AI G I LV +E G + K+ A L L
Sbjct: 156 -KMGTPTAKENAACALLRLSQIEENKVAIGRSGAIPLLVNLLETGGFRAKKDASTALYSL 214
Query: 654 C----------------------------------------VDSVRNRGLLVREGGIPPL 595
C + ++ +V EGG+P L
Sbjct: 215 CSAKENKIRAVQSGIMKPLVELMADFGSNMVDKSAFVMSLLMSVPESKPAIVEEGGVPVL 274
Query: 594 VSLSQSGTPRAKHKAETLLRYLRE 523
V + + GT R K A ++L L E
Sbjct: 275 VEIVEVGTQRQKEMAVSILLQLCE 298
Score = 37.7 bits (86), Expect = 0.31
Identities = 24/85 (28%), Positives = 38/85 (44%)
Frame = -2
Query: 750 IVEEGGIAALVEAIEDGSVKGKEFAVLTLLQLCVDSVRNRGLLVREGGIPPLVSLSQSGT 571
I + G I L+ I ++ +E+ V +L L + N+ + G I PLV + GT
Sbjct: 101 IAKAGAIKPLISLISSSDLQLQEYGVTAILNLSLCD-ENKESIASSGAIKPLVRALKMGT 159
Query: 570 PRAKHKAETLLRYLRESRQEASSSG 496
P AK A L L + + + G
Sbjct: 160 PTAKENAACALLRLSQIEENKVAIG 184
>gb|AAL16172.1|AF428404_1 AT3g01400/T13O15_4 [Arabidopsis thaliana] gi|21928049|gb|AAM78053.1|
AT3g01400/T13O15_4 [Arabidopsis thaliana]
Length = 355
Score = 196 bits (497), Expect = 7e-49
Identities = 106/194 (54%), Positives = 141/194 (72%)
Frame = -2
Query: 1062 TSKQNAACALLSLALVEENRSSIGASGAIPPLVSLLINGSSRGKKDALTTLYKLCSVKLN 883
T+K+NAACALL L+ +EEN+ +IG SGAIP LV+LL G R KKDA T LY LCS K N
Sbjct: 161 TAKENAACALLRLSQIEENKVAIGRSGAIPLLVNLLETGGFRAKKDASTALYSLCSAKEN 220
Query: 882 KERAVNAGVVKPLVDLVAEQGTGLAEKAMVVLNSLAAVQDGKDAIVEEGGIAALVEAIED 703
K RAV +G++KPLV+L+A+ G+ + +K+ V++ L +V + K AIVEEGG+ LVE +E
Sbjct: 221 KIRAVQSGIMKPLVELMADFGSNMVDKSAFVMSLLMSVPESKPAIVEEGGVPVLVEIVEV 280
Query: 702 GSVKGKEFAVLTLLQLCVDSVRNRGLLVREGGIPPLVSLSQSGTPRAKHKAETLLRYLRE 523
G+ + KE AV LLQLC +SV R ++ REG IPPLV+LSQ+GT RAK KAE L+ LR+
Sbjct: 281 GTQRQKEMAVSILLQLCEESVVYRTMVAREGAIPPLVALSQAGTSRAKQKAEALIELLRQ 340
Query: 522 SRQEASSSGS*RSN 481
R + S+G RS+
Sbjct: 341 LR--SISNGGARSS 352
Score = 70.5 bits (171), Expect = 4e-11
Identities = 57/204 (27%), Positives = 87/204 (41%), Gaps = 41/204 (20%)
Frame = -2
Query: 1011 ENRSSIGASGAIPPLVSLLINGSSRGKKDALTTLYKLCSVKLNKERAVNAGVVKPLVDLV 832
ENR I +GAI PL+SL+ + + ++ +T + L NKE ++G +KPLV +
Sbjct: 96 ENRIKIAKAGAIKPLISLISSSDLQLQEYGVTAILNLSLCDENKESIASSGAIKPLVRAL 155
Query: 831 AEQGTGLA-EKAMVVLNSLAAVQDGKDAIVEEGGIAALVEAIEDGSVKGKEFAVLTLLQL 655
+ GT A E A L L+ +++ K AI G I LV +E G + K+ A L L
Sbjct: 156 -KMGTPTAKENAACALLRLSQIEENKVAIGRSGAIPLLVNLLETGGFRAKKDASTALYSL 214
Query: 654 C----------------------------------------VDSVRNRGLLVREGGIPPL 595
C + ++ +V EGG+P L
Sbjct: 215 CSAKENKIRAVQSGIMKPLVELMADFGSNMVDKSAFVMSLLMSVPESKPAIVEEGGVPVL 274
Query: 594 VSLSQSGTPRAKHKAETLLRYLRE 523
V + + GT R K A ++L L E
Sbjct: 275 VEIVEVGTQRQKEMAVSILLQLCE 298
Score = 37.7 bits (86), Expect = 0.31
Identities = 24/85 (28%), Positives = 38/85 (44%)
Frame = -2
Query: 750 IVEEGGIAALVEAIEDGSVKGKEFAVLTLLQLCVDSVRNRGLLVREGGIPPLVSLSQSGT 571
I + G I L+ I ++ +E+ V +L L + N+ + G I PLV + GT
Sbjct: 101 IAKAGAIKPLISLISSSDLQLQEYGVTAILNLSLCD-ENKESIASSGAIKPLVRALKMGT 159
Query: 570 PRAKHKAETLLRYLRESRQEASSSG 496
P AK A L L + + + G
Sbjct: 160 PTAKENAACALLRLSQIEENKVAIG 184
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 957,817,761
Number of Sequences: 1393205
Number of extensions: 23567146
Number of successful extensions: 237668
Number of sequences better than 10.0: 4103
Number of HSP's better than 10.0 without gapping: 105501
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 175411
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 63464320210
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)