Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003467A_C01 KMC003467A_c01
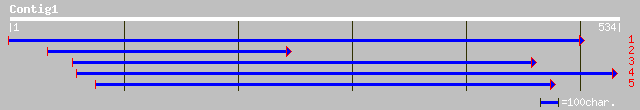
(527 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_568152.1| expressed protein; protein id: At5g05250.1, sup... 117 6e-26
ref|NP_191195.1| putative protein; protein id: At3g56360.1, supp... 99 4e-20
pir||T50848 hypothetical protein 110K5.12 [imported] - sorghum ... 59 5e-08
dbj|BAC57890.1| pherophorin [Volvox carteri f. nagariensis] 54 9e-07
sp|P21997|SSGP_VOLCA SULFATED SURFACE GLYCOPROTEIN 185 (SSG 185)... 53 3e-06
>ref|NP_568152.1| expressed protein; protein id: At5g05250.1, supported by cDNA:
gi_15810044 [Arabidopsis thaliana]
gi|10176737|dbj|BAB09968.1| gene_id:K18I23.5~unknown
protein [Arabidopsis thaliana]
gi|15810045|gb|AAL06949.1| AT5g05250/K18I23_5
[Arabidopsis thaliana]
Length = 239
Score = 117 bits (294), Expect = 6e-26
Identities = 57/104 (54%), Positives = 74/104 (70%), Gaps = 6/104 (5%)
Frame = -1
Query: 527 VKERWEAGEAPMPEGVIFMEELDEEEAAASTERT----TRVWGVVVQGKGVGSGPVCYLL 360
VK RWE GEA MPEG++F+E+L E + TR WG+VVQG+GV GPVCYLL
Sbjct: 134 VKGRWERGEAEMPEGIVFVEQLGAAEESCGCGFDGGDGTRAWGLVVQGRGVECGPVCYLL 193
Query: 359 KTSQ--AGSGPGPGMGILSTHSCLIRVKSFRESARSQLKNCWLL 234
KT++ +GSG G G+G+ TH CL +V SFRE++ SQL+NCWL+
Sbjct: 194 KTTRVGSGSGSGSGLGMRCTHFCLAKVSSFRETSESQLRNCWLV 237
>ref|NP_191195.1| putative protein; protein id: At3g56360.1, supported by cDNA:
142358. [Arabidopsis thaliana] gi|11288822|pir||T49037
hypothetical protein T5P19.10 - Arabidopsis thaliana
gi|7594514|emb|CAB88039.1| putative protein [Arabidopsis
thaliana] gi|21539571|gb|AAM53338.1| putative protein
[Arabidopsis thaliana] gi|21553412|gb|AAM62505.1|
unknown [Arabidopsis thaliana]
Length = 233
Score = 98.6 bits (244), Expect = 4e-20
Identities = 49/104 (47%), Positives = 66/104 (63%), Gaps = 7/104 (6%)
Frame = -1
Query: 518 RWEAGEAPMPEGVIFMEEL-------DEEEAAASTERTTRVWGVVVQGKGVGSGPVCYLL 360
+W GE PEGVI +E+L D++ + T WG+V QG+G +GPVCYLL
Sbjct: 132 KWGRGEERTPEGVILVEKLADGDIADDDDHDGGACGEDTSAWGIVAQGRGSDTGPVCYLL 191
Query: 359 KTSQAGSGPGPGMGILSTHSCLIRVKSFRESARSQLKNCWLLQS 228
KT++ SG MG + TH CL++VKSFRE+A SQL N WL+Q+
Sbjct: 192 KTTRVRSG----MGTVCTHFCLVKVKSFRETAMSQLNNSWLVQT 231
>pir||T50848 hypothetical protein 110K5.12 [imported] - sorghum (fragment)
gi|5410348|gb|AAD43042.1|AF124045_1 unknown [Sorghum
bicolor]
Length = 345
Score = 58.5 bits (140), Expect = 5e-08
Identities = 31/95 (32%), Positives = 49/95 (50%)
Frame = -1
Query: 527 VKERWEAGEAPMPEGVIFMEELDEEEAAASTERTTRVWGVVVQGKGVGSGPVCYLLKTSQ 348
V+ RW G P+G+I +E++ + A+++E +VWG+VVQ +G+ CY+L T +
Sbjct: 184 VRRRWGPGGQRKPDGIILVEQVAADGGASASEAARQVWGLVVQARGMECAS-CYVLDTCR 242
Query: 347 AGSGPGPGMGILSTHSCLIRVKSFRESARSQLKNC 243
S G TH CL RV+ F + C
Sbjct: 243 VRSPAG-----FCTHFCLARVQCFGDPVELPAPQC 272
>dbj|BAC57890.1| pherophorin [Volvox carteri f. nagariensis]
Length = 606
Score = 54.3 bits (129), Expect = 9e-07
Identities = 23/41 (56%), Positives = 26/41 (63%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSPS 526
P PP P S P PPPPP P+PP PPP+ +P PP SPS
Sbjct: 232 PPPPPPPPSPPPPPPPPPPPSPPPPPPPPSPSPPPPPPSPS 272
Score = 51.2 bits (121), Expect = 7e-06
Identities = 22/46 (47%), Positives = 26/46 (55%)
Frame = +2
Query: 386 QHLSLEPQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSP 523
Q++ +EPQPP P P PPPP P PP PPP P PP+ P
Sbjct: 197 QNIVVEPQPPPPPPPPPPPSPPPPPPPPPPPSPPPPPPPPPPPSPP 242
Score = 50.8 bits (120), Expect = 1e-05
Identities = 22/40 (55%), Positives = 22/40 (55%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSP 523
P PP PS P PPPPP P PP PPP S P PP P
Sbjct: 210 PPPPPPSPPPPPPPPPPPSPPPPPPPPPPPSPPPPPPPPP 249
Score = 50.4 bits (119), Expect = 1e-05
Identities = 22/41 (53%), Positives = 25/41 (60%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSPS 526
P PP P SP PPPPP P+PP PPP+ +P PP PS
Sbjct: 243 PPPPPPPPPSPPPPPPPPSPSPP--PPPPSPSPPPPPPPPS 281
Score = 49.7 bits (117), Expect = 2e-05
Identities = 24/42 (57%), Positives = 24/42 (57%), Gaps = 1/42 (2%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPAS-APHRPPTSPS 526
P PP PS P PPPPP P PP PPP S P PP SPS
Sbjct: 222 PPPPPPSPPPPPPPPPPPSPPPPPPPPPPPSPPPPPPPPSPS 263
Score = 49.3 bits (116), Expect = 3e-05
Identities = 21/38 (55%), Positives = 23/38 (60%)
Frame = +2
Query: 410 PPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSP 523
PP P SPS PPPPP P+PP PPP+ P PP P
Sbjct: 254 PPPPPPPSPSPPPPPPSPSPPPPPPPPSPPPAPPPFVP 291
Score = 47.8 bits (112), Expect = 8e-05
Identities = 21/41 (51%), Positives = 22/41 (53%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSPS 526
P PP P S P PPPPP PP PPP +P PP PS
Sbjct: 221 PPPPPPPSPPPPPPPPPPPSPPPPPPPPPPPSPPPPPPPPS 261
Score = 47.0 bits (110), Expect = 1e-04
Identities = 21/37 (56%), Positives = 21/37 (56%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPP 514
P PP P S SP PPP P P PP PPP S P PP
Sbjct: 254 PPPPPPPSPSPPPPPPSPSPPPP---PPPPSPPPAPP 287
Score = 47.0 bits (110), Expect = 1e-04
Identities = 21/40 (52%), Positives = 22/40 (54%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSP 523
P PP P PS PPPPP P PP PPP P PP+ P
Sbjct: 217 PPPPPPPPPPPSPPPPPPPPPPP--SPPPPPPPPPPPSPP 254
Score = 44.3 bits (103), Expect = 9e-04
Identities = 21/43 (48%), Positives = 22/43 (50%), Gaps = 3/43 (6%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPP---PHPTPP*I*PPPASAPHRPPTSP 523
P PP P S P PPPP P P PP PPP P PP +P
Sbjct: 244 PPPPPPPPSPPPPPPPPSPSPPPPPPSPSPPPPPPPPSPPPAP 286
Score = 40.0 bits (92), Expect = 0.017
Identities = 20/37 (54%), Positives = 20/37 (54%), Gaps = 1/37 (2%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPP-PHPTPP*I*PPPASAPHRP 511
P PP PS S P PP P P P PP PPPA P P
Sbjct: 255 PPPPPPSPSPPPPPPSPSPPPPPPPPSPPPAPPPFVP 291
Score = 38.1 bits (87), Expect = 0.064
Identities = 18/31 (58%), Positives = 19/31 (61%)
Frame = +2
Query: 395 SLEPQPPTPSSSSPSTPPPPPHPTPP*I*PP 487
S P PP P SPS PPPPP P+PP PP
Sbjct: 261 SPSPPPPPP---SPSPPPPPPPPSPPPAPPP 288
>sp|P21997|SSGP_VOLCA SULFATED SURFACE GLYCOPROTEIN 185 (SSG 185) gi|99441|pir||A33647
sulfated surface glycoprotein 185 - Volvox carteri
gi|1405821|emb|CAA35953.1| SULFATED SURFACE GLYCOPROTEIN
185 [Volvox carteri]
Length = 485
Score = 52.8 bits (125), Expect = 3e-06
Identities = 22/40 (55%), Positives = 25/40 (62%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSP 523
P+PP+P SPS PPPPP P PP PPP+ P PP P
Sbjct: 248 PRPPSPPPPSPSPPPPPPPPPPPPPPPPPSPPPPPPPPPP 287
Score = 50.8 bits (120), Expect = 1e-05
Identities = 23/41 (56%), Positives = 25/41 (60%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSPS 526
P PP P S P PPPPP P PP PPP+ +P R P SPS
Sbjct: 269 PPPPPPPPSPPPPPPPPPPPPPP--PPPPSPSPPRKPPSPS 307
Score = 49.7 bits (117), Expect = 2e-05
Identities = 21/40 (52%), Positives = 22/40 (54%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSP 523
P PP+PS P PPPPP P PP PPP P PP P
Sbjct: 253 PPPPSPSPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPP 292
Score = 48.9 bits (115), Expect = 4e-05
Identities = 26/62 (41%), Positives = 29/62 (45%)
Frame = +2
Query: 341 TRPGSSSVNNKPDRTQHLSLEPQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTS 520
+RP S + +P S P PP P P PPPP P PP PPP P PP S
Sbjct: 239 SRPPSPPPSPRPPSPPPPSPSPPPPPPPPPPPPPPPPPSPPPPP--PPPPPPPPPPPPPS 296
Query: 521 PS 526
PS
Sbjct: 297 PS 298
Score = 45.1 bits (105), Expect = 5e-04
Identities = 21/40 (52%), Positives = 22/40 (54%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTSP 523
P PP P P PPPPP P+PP PP S P PP SP
Sbjct: 278 PPPPPPPPPPPPPPPPPPSPSPP-RKPPSPSPPVPPPPSP 316
Score = 45.1 bits (105), Expect = 5e-04
Identities = 23/68 (33%), Positives = 29/68 (41%)
Frame = +2
Query: 320 PFLDRVQTRPGSSSVNNKPDRTQHLSLEPQPPTPSSSSPSTPPPPPHPTPP*I*PPPASA 499
P ++ + P +S + P T PP+P SP P P P P PP PPP
Sbjct: 215 PNVNPIGPAPNNSPLPPSPQPTASSRPPSPPPSPRPPSPPPPSPSPPPPPPPPPPPPPPP 274
Query: 500 PHRPPTSP 523
P PP P
Sbjct: 275 PPSPPPPP 282
Score = 39.3 bits (90), Expect = 0.029
Identities = 19/39 (48%), Positives = 21/39 (53%)
Frame = +2
Query: 404 PQPPTPSSSSPSTPPPPPHPTPP*I*PPPASAPHRPPTS 520
P PP P SPS P PP P+PP PPP S P P +
Sbjct: 287 PPPPPPPPPSPSPPRKPPSPSPPV--PPPPSPPSVLPAA 323
Score = 33.1 bits (74), Expect = 2.1
Identities = 16/38 (42%), Positives = 19/38 (49%), Gaps = 5/38 (13%)
Frame = +2
Query: 404 PQPPTPSSSSPS-----TPPPPPHPTPP*I*PPPASAP 502
P PP PS S P +PP PP P+PP + P P
Sbjct: 290 PPPPPPSPSPPRKPPSPSPPVPPPPSPPSVLPAATGFP 327
Score = 32.0 bits (71), Expect = 4.6
Identities = 25/79 (31%), Positives = 28/79 (34%), Gaps = 5/79 (6%)
Frame = +2
Query: 305 GSTIYPFLDRVQTRPGSSSVNNKPDRTQHLSLEPQPPTPSSSSPSTPPPPPHPTP----- 469
GS +P L S SV N L P +P+ P PP P P
Sbjct: 181 GSCTFPELFMDVNGTASYSVFNSDKDCCPTGLSGPNVNPIGPAPNNSPLPPSPQPTASSR 240
Query: 470 P*I*PPPASAPHRPPTSPS 526
P PP P PP SPS
Sbjct: 241 PPSPPPSPRPPSPPPPSPS 259
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 519,860,796
Number of Sequences: 1393205
Number of extensions: 14894784
Number of successful extensions: 297711
Number of sequences better than 10.0: 4667
Number of HSP's better than 10.0 without gapping: 88348
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 196382
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17308240320
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)