Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003459A_C02 KMC003459A_c02
(598 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
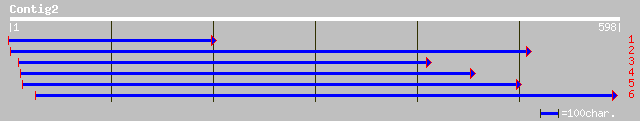
ref|NP_566091.1| Expressed protein; protein id: At2g46910.1, sup... 186 3e-46
gb|AAM62945.1| unknown [Arabidopsis thaliana] 186 3e-46
gb|AAM94938.1| unknown protein [Oryza sativa (japonica cultivar-... 93 7e-20
>ref|NP_566091.1| Expressed protein; protein id: At2g46910.1, supported by cDNA:
17422., supported by cDNA: gi_17065041, supported by
cDNA: gi_20259993 [Arabidopsis thaliana]
gi|17065042|gb|AAL32675.1| Unknown protein [Arabidopsis
thaliana] gi|20197139|gb|AAC34229.2| Expressed protein
[Arabidopsis thaliana] gi|20259994|gb|AAM13344.1|
unknown protein [Arabidopsis thaliana]
Length = 284
Score = 186 bits (471), Expect = 3e-46
Identities = 92/121 (76%), Positives = 111/121 (91%), Gaps = 2/121 (1%)
Frame = -3
Query: 596 WSVPNLLEEQDGATLLVSAKFTVVSVRNTYLQFQEITLQDININEQLQALIAPALLPRSY 417
WS+P+LLEEQ+GATL+V+AKF VS RN YLQF+EI++++ININEQLQALIAPA+LPRS+
Sbjct: 162 WSLPSLLEEQEGATLVVTAKFDKVSSRNIYLQFEEISVRNININEQLQALIAPAILPRSF 221
Query: 416 LSLQILQFLRAFKAQIPVR--DPGRQSVGGLYYLSYLDDNMLLGRAVGGGGVFVFTRAQS 243
LSLQ+LQF+R FKAQIPV PGR+SVGGLYYLSYLD+NMLLGR+VGGGGVFVFT++Q
Sbjct: 222 LSLQLLQFIRTFKAQIPVNATSPGRRSVGGLYYLSYLDNNMLLGRSVGGGGVFVFTKSQP 281
Query: 242 L 240
L
Sbjct: 282 L 282
>gb|AAM62945.1| unknown [Arabidopsis thaliana]
Length = 284
Score = 186 bits (471), Expect = 3e-46
Identities = 92/121 (76%), Positives = 111/121 (91%), Gaps = 2/121 (1%)
Frame = -3
Query: 596 WSVPNLLEEQDGATLLVSAKFTVVSVRNTYLQFQEITLQDININEQLQALIAPALLPRSY 417
WS+P+LLEEQ+GATL+V+AKF VS RN YLQF+EI++++ININEQLQALIAPA+LPRS+
Sbjct: 162 WSLPSLLEEQEGATLVVTAKFDKVSSRNIYLQFEEISVRNININEQLQALIAPAILPRSF 221
Query: 416 LSLQILQFLRAFKAQIPVR--DPGRQSVGGLYYLSYLDDNMLLGRAVGGGGVFVFTRAQS 243
LSLQ+LQF+R FKAQIPV PGR+SVGGLYYLSYLD+NMLLGR+VGGGGVFVFT++Q
Sbjct: 222 LSLQLLQFIRTFKAQIPVNATSPGRRSVGGLYYLSYLDNNMLLGRSVGGGGVFVFTKSQP 281
Query: 242 L 240
L
Sbjct: 282 L 282
>gb|AAM94938.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 126
Score = 92.8 bits (229), Expect(2) = 7e-20
Identities = 43/66 (65%), Positives = 57/66 (86%)
Frame = -3
Query: 551 LVSAKFTVVSVRNTYLQFQEITLQDININEQLQALIAPALLPRSYLSLQILQFLRAFKAQ 372
+VSAKF V+S RN +LQF+E+ +++I I+EQLQALIAPA+LPRS+ SLQILQFL+ F+AQ
Sbjct: 1 MVSAKFAVMSKRNIFLQFEEVVVENIKISEQLQALIAPAILPRSFFSLQILQFLKTFRAQ 60
Query: 371 IPVRDP 354
+PV P
Sbjct: 61 VPVNGP 66
Score = 26.2 bits (56), Expect(2) = 7e-20
Identities = 15/42 (35%), Positives = 20/42 (46%)
Frame = -1
Query: 349 GNR*EDYITLVIWMITCFWVVLLVEEGYLCLREPSRSTDEHR 224
G+ EDYI + C W LV E +L +E S D+ R
Sbjct: 74 GDHPEDYIIFLTLTAICSWGDRLVVEEFLFSQEHSLFCDDFR 115
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 478,348,727
Number of Sequences: 1393205
Number of extensions: 9831880
Number of successful extensions: 23496
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 22557
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23448
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23140425222
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)