Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
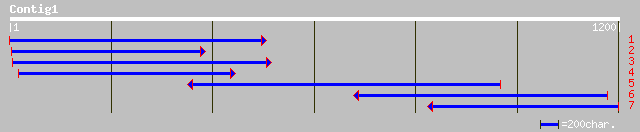
Query= KMC003450A_C01 KMC003450A_c01
(1178 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||S66262 vestitone reductase - alfalfa gi|973249|gb|AAB41550.... 525 e-148
pir||T07104 2'-hydroxydihydrodaidzein reductase - soybean gi|268... 511 e-144
gb|AAD17997.1| sophorol reductase [Pisum sativum] 490 e-137
gb|AAF17576.1|AF202182_1 2'-hydroxy isoflavone/dihydroflavonol r... 434 e-120
ref|NP_182064.1| putative flavonol reductase; protein id: At2g45... 324 2e-87
>pir||S66262 vestitone reductase - alfalfa gi|973249|gb|AAB41550.1| vestitone
reductase
Length = 326
Score = 525 bits (1351), Expect = e-148
Identities = 261/328 (79%), Positives = 294/328 (89%)
Frame = -2
Query: 1162 MAEGKGRVCVTGGTGFLASWIIKRLLEDGYSVNTTIRSNSGGKRDVSFLTNLPGATSEKL 983
MAEGKGRVCVTGGTGFL SWIIK LLE+GYSVNTTIR++ KRDVSFLTNLPGA SEKL
Sbjct: 1 MAEGKGRVCVTGGTGFLGSWIIKSLLENGYSVNTTIRADPERKRDVSFLTNLPGA-SEKL 59
Query: 982 QIFNADLCIPESFGPAVEGCVGIFHTATPVDFAVNEPEEVVTKRTVDGALGILKACVNSK 803
FNADL P+SF A+EGCVGIFHTA+P+DFAV+EPEE+VTKRTVDGALGILKACVNSK
Sbjct: 60 HFFNADLSNPDSFAAAIEGCVGIFHTASPIDFAVSEPEEIVTKRTVDGALGILKACVNSK 119
Query: 802 TVKRVVYTSSGSAASFSGKDGGDALDESYWSDVDLLRKVKPFGWSYAVSKTXAEKAVLEF 623
TVKR +YTSSGSA SF+GKD D LDES WSDVDLLR VKPFGW+YAVSKT AEKAVLEF
Sbjct: 120 TVKRFIYTSSGSAVSFNGKDK-DVLDESDWSDVDLLRSVKPFGWNYAVSKTLAEKAVLEF 178
Query: 622 GEKHGLEVVTLIPTFVVGPWVCPKLPDSVERALVLVLGKKEQIGVIRFHMVHVDDVARAH 443
GE++G++VVTLI F+VG +VCPKLPDS+E+ALVLVLGKKEQIGV RFHMVHVDDVARAH
Sbjct: 179 GEQNGIDVVTLILPFIVGRFVCPKLPDSIEKALVLVLGKKEQIGVTRFHMVHVDDVARAH 238
Query: 442 IFLLEHPNPKGRYNCSPFIVPIEEMSKLLSAKYPEYQIPSVEELKGIEGFKQPDLISNKI 263
I+LLE+ P GRYNCSPFIVPIEEMS+LLSAKYPEYQI +V+ELK I+G + PDL + K+
Sbjct: 239 IYLLENSVPGGRYNCSPFIVPIEEMSQLLSAKYPEYQILTVDELKEIKGARLPDLNTKKL 298
Query: 262 RDAGFEFKYSLEDLFDDAIQCCKEKGYL 179
DAGF+FKY++ED+FDDAIQCCKEKGYL
Sbjct: 299 VDAGFDFKYTIEDMFDDAIQCCKEKGYL 326
>pir||T07104 2'-hydroxydihydrodaidzein reductase - soybean
gi|2687726|emb|CAA06028.1| 2'-hydroxydihydrodaidzein
reductase [Glycine max]
Length = 327
Score = 511 bits (1316), Expect = e-144
Identities = 253/328 (77%), Positives = 284/328 (86%)
Frame = -2
Query: 1162 MAEGKGRVCVTGGTGFLASWIIKRLLEDGYSVNTTIRSNSGGKRDVSFLTNLPGATSEKL 983
M EGKGR+CVTGGTGFL SWIIK LLE GY+VNTTIRS+ G KRDVSFLTNLPGA SEKL
Sbjct: 1 MGEGKGRICVTGGTGFLGSWIIKSLLEHGYAVNTTIRSDPGRKRDVSFLTNLPGA-SEKL 59
Query: 982 QIFNADLCIPESFGPAVEGCVGIFHTATPVDFAVNEPEEVVTKRTVDGALGILKACVNSK 803
+IFNADL PESFGPAVEGCVGIFHTATP+DFAVNEPEEVVTKR +DGALGILKA + +K
Sbjct: 60 KIFNADLSDPESFGPAVEGCVGIFHTATPIDFAVNEPEEVVTKRAIDGALGILKAGLKAK 119
Query: 802 TVKRVVYTSSGSAASFSGKDGGDALDESYWSDVDLLRKVKPFGWSYAVSKTXAEKAVLEF 623
TVKRVVYTSS S SFS + D +DES WSDVDLLR VKPF WSYAVSK +EKAVLEF
Sbjct: 120 TVKRVVYTSSASTVSFSSLEEKDVVDESVWSDVDLLRSVKPFSWSYAVSKVLSEKAVLEF 179
Query: 622 GEKHGLEVVTLIPTFVVGPWVCPKLPDSVERALVLVLGKKEQIGVIRFHMVHVDDVARAH 443
GE++GLEV TL+ FVVG +VCPKLPDSVERAL+LVLGKKE+IGVIR+HMVHVDDVARAH
Sbjct: 180 GEQNGLEVTTLVLPFVVGRFVCPKLPDSVERALLLVLGKKEEIGVIRYHMVHVDDVARAH 239
Query: 442 IFLLEHPNPKGRYNCSPFIVPIEEMSKLLSAKYPEYQIPSVEELKGIEGFKQPDLISNKI 263
IFLLEHPNPKGRYNCSPFIVPIEE+++++SAKYPEYQIP++EE+K I+G K P L S K+
Sbjct: 240 IFLLEHPNPKGRYNCSPFIVPIEEIAEIISAKYPEYQIPALEEVKEIKGAKLPHLTSQKL 299
Query: 262 RDAGFEFKYSLEDLFDDAIQCCKEKGYL 179
DA +ED+F DAI+CCKEKGYL
Sbjct: 300 VDAVLSSSICVEDIFTDAIECCKEKGYL 327
>gb|AAD17997.1| sophorol reductase [Pisum sativum]
Length = 326
Score = 490 bits (1261), Expect = e-137
Identities = 242/328 (73%), Positives = 287/328 (86%)
Frame = -2
Query: 1162 MAEGKGRVCVTGGTGFLASWIIKRLLEDGYSVNTTIRSNSGGKRDVSFLTNLPGATSEKL 983
MAEGKGRVCVTGGTGFL SWIIK LLE+GYSVNTT+R + KRD+SFLTNLPGA SE+L
Sbjct: 1 MAEGKGRVCVTGGTGFLGSWIIKSLLENGYSVNTTVRPDPKRKRDLSFLTNLPGA-SERL 59
Query: 982 QIFNADLCIPESFGPAVEGCVGIFHTATPVDFAVNEPEEVVTKRTVDGALGILKACVNSK 803
FNADL P+SF AV+GCVG+FHTATPV+FAV+EPE++VTKRTVDGALGILKACVNSK
Sbjct: 60 HFFNADLSDPKSFSAAVKGCVGVFHTATPVNFAVSEPEKIVTKRTVDGALGILKACVNSK 119
Query: 802 TVKRVVYTSSGSAASFSGKDGGDALDESYWSDVDLLRKVKPFGWSYAVSKTXAEKAVLEF 623
T+KR +YTSSGSA SF+GK+ + LDE+ WSDV LL+ VKPFGWSY+VSKT AEKAVL+F
Sbjct: 120 TLKRFIYTSSGSAVSFNGKNK-EVLDETDWSDVGLLKSVKPFGWSYSVSKTLAEKAVLDF 178
Query: 622 GEKHGLEVVTLIPTFVVGPWVCPKLPDSVERALVLVLGKKEQIGVIRFHMVHVDDVARAH 443
G+++G++V TLI F+VG ++CPKLPDSV++ALVLVLGKKEQIGV RFHMVHVDDVARAH
Sbjct: 179 GQQNGIDVATLILPFIVGSFICPKLPDSVDKALVLVLGKKEQIGVTRFHMVHVDDVARAH 238
Query: 442 IFLLEHPNPKGRYNCSPFIVPIEEMSKLLSAKYPEYQIPSVEELKGIEGFKQPDLISNKI 263
I+LLE+ P GRYNC+PFIV IEEMS+L SAKYPE+QI SV+ELK I+G + +L S K+
Sbjct: 239 IYLLENSVPGGRYNCAPFIVTIEEMSQLFSAKYPEFQILSVDELKEIKGARLAELNSKKL 298
Query: 262 RDAGFEFKYSLEDLFDDAIQCCKEKGYL 179
AGFEFKYS++D+F+DAIQCCKEKGYL
Sbjct: 299 VAAGFEFKYSVDDMFEDAIQCCKEKGYL 326
>gb|AAF17576.1|AF202182_1 2'-hydroxy isoflavone/dihydroflavonol reductase homolog [Glycine max]
Length = 326
Score = 434 bits (1116), Expect = e-120
Identities = 219/330 (66%), Positives = 261/330 (78%), Gaps = 1/330 (0%)
Frame = -2
Query: 1165 QMAEGKGRVCVTGGTGFLASWIIKRLLEDGYSVNTTIRSNSGGKRDVSFLTNLPGATSEK 986
+ KGRVCVTGGTGF+ SWIIKRLLEDGYSVNTT+R + ++DVSFLT+LP A S++
Sbjct: 1 EFGTSKGRVCVTGGTGFIGSWIIKRLLEDGYSVNTTVRPDPEHRKDVSFLTSLPRA-SQR 59
Query: 985 LQIFNADLCIPESFGPAVEGCVGIFHTATPVDFAVNEPEEVVTKRTVDGALGILKACVNS 806
LQI +ADL PESF ++EGC+G+FH ATPVDF + EPEEVVTKR+++GALGILKAC+NS
Sbjct: 60 LQILSADLSNPESFIASIEGCMGVFHVATPVDFELREPEEVVTKRSIEGALGILKACLNS 119
Query: 805 KTVKRVVYTSSGSAASFSGKDGGDALDESYWSDVDLLRKVKPFGWSYAVSKTXAEKAVLE 626
KTVKRVVYTSS SA + ++ +DES W+DVD LR KPFGWSY+VSKT EKAVLE
Sbjct: 120 KTVKRVVYTSSASAVDNNKEE---IMDESSWNDVDYLRSSKPFGWSYSVSKTLTEKAVLE 176
Query: 625 FGEKHGLEVVTLIPTFVVGPWVCPKLPDSVERALVLVLGKKEQIGVI-RFHMVHVDDVAR 449
FGE++GL+VVTLIPT V GP++CPKLP SV +L +LG+K GV+ + MVHVDDVAR
Sbjct: 177 FGEQNGLDVVTLIPTLVFGPFICPKLPSSVRNSLDFILGEKGTFGVVLQTDMVHVDDVAR 236
Query: 448 AHIFLLEHPNPKGRYNCSPFIVPIEEMSKLLSAKYPEYQIPSVEELKGIEGFKQPDLISN 269
AHIFLLEHPNPKGRY CS V E +SKL+SAKYPE+Q P VE L IEG K L S
Sbjct: 237 AHIFLLEHPNPKGRYICSQCSVTYERISKLVSAKYPEFQPPPVESLNHIEGTKGSYLSSK 296
Query: 268 KIRDAGFEFKYSLEDLFDDAIQCCKEKGYL 179
K+ DAGF +KY LE++ DDAIQCCKEKGYL
Sbjct: 297 KLIDAGFVYKYGLEEMVDDAIQCCKEKGYL 326
>ref|NP_182064.1| putative flavonol reductase; protein id: At2g45400.1 [Arabidopsis
thaliana] gi|25284025|pir||A84890 probable flavonol
reductase [imported] - Arabidopsis thaliana
gi|2583115|gb|AAB82624.1| putative flavonol reductase
[Arabidopsis thaliana]
Length = 364
Score = 324 bits (830), Expect = 2e-87
Identities = 177/339 (52%), Positives = 233/339 (68%), Gaps = 6/339 (1%)
Frame = -2
Query: 1177 EECVQMAEGKGRVCVTGGTGFLASWIIKRLLEDGYSVNTTIRSNS-GGKRDVSFLTNLPG 1001
E + + G VCVTGG+GF+ASW+I RLL+ GYSV T+R+NS G K+D+S+LT LP
Sbjct: 28 ETVPSLLDETGLVCVTGGSGFVASWLIMRLLQRGYSVRATVRTNSEGNKKDISYLTELPF 87
Query: 1000 ATSEKLQIFNADLCIPESFGPAVEGCVGIFHTATPVDFAVNEPEEVVTKRTVDGALGILK 821
A SE+LQIF ADL PESF PA+EGC +FH A P+D NE EE VTKRTV G +GILK
Sbjct: 88 A-SERLQIFTADLNEPESFKPAIEGCKAVFHVAHPMDPNSNETEETVTKRTVQGLMGILK 146
Query: 820 ACVNSKTVKRVVYTSSGSAASFSGKDGGDA--LDESYWSDVDLLR--KVKPFGWSYAVSK 653
+C+++KTVKR YTSS +SG +GG +DES WSDV++ R K K SY VSK
Sbjct: 147 SCLDAKTVKRFFYTSSAVTVFYSGGNGGGGGEVDESVWSDVEVFRNQKEKRVSSSYVVSK 206
Query: 652 TXAEKAVLEFGEKHGLEVVTLIPTFVVGPWVCPKLPDSVERALVLVLGK-KEQIGVIRFH 476
AE A LEFG K+GLEVVTL+ VVGP++ LP SV +L ++ G KE+ ++
Sbjct: 207 MAAETAALEFGGKNGLEVVTLVIPLVVGPFISSSLPSSVFISLAMLFGNYKEKYLFDTYN 266
Query: 475 MVHVDDVARAHIFLLEHPNPKGRYNCSPFIVPIEEMSKLLSAKYPEYQIPSVEELKGIEG 296
MVH+DDVARA IFLLE P KGRY CS + I+E+ + LS K+P++Q+PS+ +L +
Sbjct: 267 MVHIDDVARAMIFLLEKPVAKGRYICSSVEMKIDEVFEFLSTKFPQFQLPSI-DLNKYKV 325
Query: 295 FKQPDLISNKIRDAGFEFKYSLEDLFDDAIQCCKEKGYL 179
K+ L S K++ AGFEFKY E++F AI+ C+ +G+L
Sbjct: 326 EKRMGLSSKKLKSAGFEFKYGAEEIFSGAIRSCQARGFL 364
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,016,240,787
Number of Sequences: 1393205
Number of extensions: 22944995
Number of successful extensions: 72125
Number of sequences better than 10.0: 894
Number of HSP's better than 10.0 without gapping: 66082
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 70950
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 73301812074
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)