Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003449A_C01 KMC003449A_c01
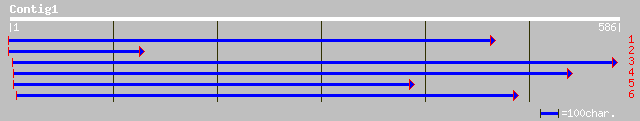
(586 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T00886 hypothetical protein F17K2.26 - Arabidopsis thaliana 174 6e-43
ref|NP_566054.1| expressed protein; protein id: At2g45730.1, sup... 174 6e-43
emb|CAD39377.1| OSJNBb0021I10.12 [Oryza sativa (japonica cultiva... 161 6e-39
ref|NP_057023.2| CGI-09 protein [Homo sapiens] gi|5834567|emb|CA... 74 1e-12
ref|XP_230548.1| similar to expressed sequence AW492787 [Mus mus... 74 1e-12
>pir||T00886 hypothetical protein F17K2.26 - Arabidopsis thaliana
Length = 335
Score = 174 bits (442), Expect = 6e-43
Identities = 81/126 (64%), Positives = 102/126 (80%)
Frame = -2
Query: 552 GADNAITPATKVGKATKAGDKAPQEIIDLWKENGFSSLIIAAPELDTWTLVRDLFPLLSN 373
G + + P +K+ KA KAG KA +E I++W+ENGFSSLI+AA + D W + +D+ PLLS
Sbjct: 207 GVVDIVVPESKIIKAAKAGAKASKETIEMWQENGFSSLIMAAQDQDPWIVAKDVLPLLSY 266
Query: 372 SAPFAIYHQHLQPLATCMRNLQLEKMAVGLQLSEPWLREYQVLPSRTHPCMQMSAFGGYI 193
SAPFAIYHQ+LQPLATCM LQ KMA+ LQ++EPWLREYQVLPSR+HP MQMS+FGGY+
Sbjct: 267 SAPFAIYHQYLQPLATCMHKLQQGKMAINLQITEPWLREYQVLPSRSHPHMQMSSFGGYV 326
Query: 192 LSGTKI 175
LSG +I
Sbjct: 327 LSGIRI 332
>ref|NP_566054.1| expressed protein; protein id: At2g45730.1, supported by cDNA:
9000., supported by cDNA: gi_17473623 [Arabidopsis
thaliana] gi|7486467|pir||T02474 hypothetical protein
At2g45730 [imported] - Arabidopsis thaliana
gi|3386622|gb|AAC28552.1| expressed protein [Arabidopsis
thaliana] gi|17473624|gb|AAL38275.1| unknown protein
[Arabidopsis thaliana] gi|22136132|gb|AAM91144.1|
unknown protein [Arabidopsis thaliana]
Length = 446
Score = 174 bits (442), Expect = 6e-43
Identities = 81/126 (64%), Positives = 102/126 (80%)
Frame = -2
Query: 552 GADNAITPATKVGKATKAGDKAPQEIIDLWKENGFSSLIIAAPELDTWTLVRDLFPLLSN 373
G + + P +K+ KA KAG KA +E I++W+ENGFSSLI+AA + D W + +D+ PLLS
Sbjct: 318 GVVDIVVPESKIIKAAKAGAKASKETIEMWQENGFSSLIMAAQDQDPWIVAKDVLPLLSY 377
Query: 372 SAPFAIYHQHLQPLATCMRNLQLEKMAVGLQLSEPWLREYQVLPSRTHPCMQMSAFGGYI 193
SAPFAIYHQ+LQPLATCM LQ KMA+ LQ++EPWLREYQVLPSR+HP MQMS+FGGY+
Sbjct: 378 SAPFAIYHQYLQPLATCMHKLQQGKMAINLQITEPWLREYQVLPSRSHPHMQMSSFGGYV 437
Query: 192 LSGTKI 175
LSG +I
Sbjct: 438 LSGIRI 443
>emb|CAD39377.1| OSJNBb0021I10.12 [Oryza sativa (japonica cultivar-group)]
Length = 475
Score = 161 bits (407), Expect = 6e-39
Identities = 79/113 (69%), Positives = 91/113 (79%)
Frame = -2
Query: 513 KATKAGDKAPQEIIDLWKENGFSSLIIAAPELDTWTLVRDLFPLLSNSAPFAIYHQHLQP 334
K+ KAG E + W E+GFSSLI+AAP D + V DL PLLS SAPFAIYHQ+LQP
Sbjct: 353 KSLKAGKAPSPERMKYWSEHGFSSLIVAAPGHDVESFVADLLPLLSYSAPFAIYHQYLQP 412
Query: 333 LATCMRNLQLEKMAVGLQLSEPWLREYQVLPSRTHPCMQMSAFGGYILSGTKI 175
LATCM +LQ+ KMA+GLQ+SEPWLREYQVLPSRTHP MQM+AFGGYILSG +I
Sbjct: 413 LATCMHSLQVSKMALGLQISEPWLREYQVLPSRTHPHMQMNAFGGYILSGIRI 465
>ref|NP_057023.2| CGI-09 protein [Homo sapiens] gi|5834567|emb|CAB55274.1|
dJ967N21.3.1 (novel protein (isoform 1)) [Homo sapiens]
gi|12654837|gb|AAH01262.1|AAH01262 CGI-09 protein [Homo
sapiens]
Length = 497
Score = 73.9 bits (180), Expect = 1e-12
Identities = 41/99 (41%), Positives = 55/99 (55%)
Frame = -2
Query: 480 EIIDLWKENGFSSLIIAAPELDTWTLVRDLFPLLSNSAPFAIYHQHLQPLATCMRNLQLE 301
E L E LI+A+ T L+ L ++ S PF +Y Q+ +PL C L+
Sbjct: 359 EAAALLSERNADGLIVASRFHPT-PLLLSLLDFVAPSRPFVVYCQYKEPLLECYTKLRER 417
Query: 300 KMAVGLQLSEPWLREYQVLPSRTHPCMQMSAFGGYILSG 184
+ L+LSE WLR YQVLP R+HP + MS GGY+LSG
Sbjct: 418 GGVINLRLSETWLRNYQVLPDRSHPKLLMSGGGGYLLSG 456
>ref|XP_230548.1| similar to expressed sequence AW492787 [Mus musculus] [Rattus
norvegicus]
Length = 504
Score = 73.9 bits (180), Expect = 1e-12
Identities = 42/103 (40%), Positives = 56/103 (53%)
Frame = -2
Query: 492 KAPQEIIDLWKENGFSSLIIAAPELDTWTLVRDLFPLLSNSAPFAIYHQHLQPLATCMRN 313
K E L E LI+A+ T L+ L ++ S PF +Y Q+ +PL C
Sbjct: 362 KRHMEAAALLGERNADGLIVASRFHPT-PLLLSLLDFVAPSRPFVVYCQYKEPLLECYTK 420
Query: 312 LQLEKMAVGLQLSEPWLREYQVLPSRTHPCMQMSAFGGYILSG 184
L+ + L+LSE WLR YQVLP R+HP + MS GGY+LSG
Sbjct: 421 LRERGGVINLRLSETWLRNYQVLPDRSHPKLLMSGGGGYLLSG 463
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 488,939,760
Number of Sequences: 1393205
Number of extensions: 10356910
Number of successful extensions: 26835
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 26274
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26830
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)