Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003448A_C01 KMC003448A_c01
(586 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
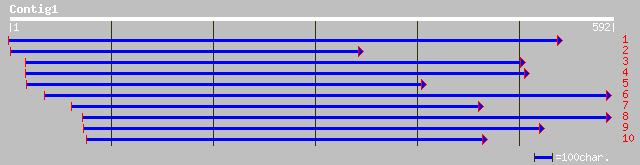
ref|NP_196563.1| hypothetical protein; protein id: At5g10010.1 [... 115 5e-25
gb|AAM28290.1| putative ATP/GTP binding protein [Ananas comosus] 107 1e-22
ref|NP_201296.1| unknown protein; protein id: At5g64910.1 [Arabi... 56 4e-07
gb|AAN17926.1| Erp45 protein [Borrelia burgdorferi] 34 1.5
gb|AAD28093.1|AF077602_2 ElpA1 protein [Borrelia burgdorferi] 34 1.5
>ref|NP_196563.1| hypothetical protein; protein id: At5g10010.1 [Arabidopsis
thaliana]
Length = 273
Score = 115 bits (287), Expect = 5e-25
Identities = 54/70 (77%), Positives = 63/70 (89%)
Frame = -1
Query: 586 EKVREAKRAXREAREARRKAIEEMSEETKAAFESMKFYKFYPVQSPDTPDLSEVKSPFIN 407
E+VR AK+A REA++AR+KAIEEMSE+TK AF+ MKFYKFYP SPDTPD+S V+SPFIN
Sbjct: 204 EQVRAAKKANREAKDARKKAIEEMSEDTKQAFQKMKFYKFYPQPSPDTPDVSGVQSPFIN 263
Query: 406 RYYGKAHEVL 377
RYYGKAHEVL
Sbjct: 264 RYYGKAHEVL 273
>gb|AAM28290.1| putative ATP/GTP binding protein [Ananas comosus]
Length = 364
Score = 107 bits (267), Expect = 1e-22
Identities = 49/70 (70%), Positives = 61/70 (87%)
Frame = -1
Query: 586 EKVREAKRAXREAREARRKAIEEMSEETKAAFESMKFYKFYPVQSPDTPDLSEVKSPFIN 407
E+VRE K+ ++AREARRKAIE+M +TKAAFE+M+FYKFYPVQ+ DTPD+S VK P+IN
Sbjct: 295 EQVRETKKTQKQAREARRKAIEDMDPKTKAAFENMQFYKFYPVQTLDTPDISSVKVPYIN 354
Query: 406 RYYGKAHEVL 377
RYYGKAH V+
Sbjct: 355 RYYGKAHVVM 364
>ref|NP_201296.1| unknown protein; protein id: At5g64910.1 [Arabidopsis thaliana]
gi|8843755|dbj|BAA97303.1| gene_id:MXK3.14~unknown
protein [Arabidopsis thaliana]
Length = 487
Score = 55.8 bits (133), Expect = 4e-07
Identities = 27/55 (49%), Positives = 37/55 (67%)
Frame = -1
Query: 586 EKVREAKRAXREAREARRKAIEEMSEETKAAFESMKFYKFYPVQSPDTPDLSEVK 422
EK +A A A+EA KA E +SEETK A++ M+ YKFYP+ SPDTP + ++
Sbjct: 404 EKSDKAMAAYDTAQEALEKAKEGLSEETKKAYQEMRLYKFYPLPSPDTPHTAGIE 458
>gb|AAN17926.1| Erp45 protein [Borrelia burgdorferi]
Length = 368
Score = 33.9 bits (76), Expect = 1.5
Identities = 16/36 (44%), Positives = 21/36 (57%)
Frame = -1
Query: 586 EKVREAKRAXREAREARRKAIEEMSEETKAAFESMK 479
EK+ E K +EA E RR+ IE+ +E KA E K
Sbjct: 160 EKIEEEKAKEKEAEELRRRRIEQYKKEEKARLERKK 195
>gb|AAD28093.1|AF077602_2 ElpA1 protein [Borrelia burgdorferi]
Length = 303
Score = 33.9 bits (76), Expect = 1.5
Identities = 16/36 (44%), Positives = 21/36 (57%)
Frame = -1
Query: 586 EKVREAKRAXREAREARRKAIEEMSEETKAAFESMK 479
EK+ E K +EA E RR+ IE+ +E KA E K
Sbjct: 160 EKIEEEKAKEKEAEELRRRRIEQYKKEEKARLERKK 195
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 471,379,658
Number of Sequences: 1393205
Number of extensions: 9799721
Number of successful extensions: 22757
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 22067
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22742
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)