Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003425A_C01 KMC003425A_c01
(710 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
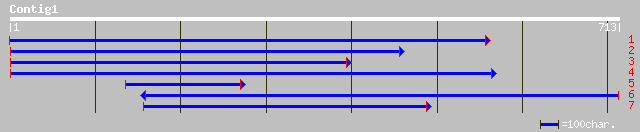
ref|NP_175926.1| Serine/arginine-rich protein, putative; protein... 171 7e-42
dbj|BAC00616.1| OJ1316_H05.12 [Oryza sativa (japonica cultivar-g... 107 1e-22
ref|NP_499647.1| TBP-Associated transcription Factor family TAF-... 60 2e-08
gb|EAA09978.1| agCP11819 [Anopheles gambiae str. PEST] 58 1e-07
gb|EAA34629.1| hypothetical protein [Neurospora crassa] 57 3e-07
>ref|NP_175926.1| Serine/arginine-rich protein, putative; protein id: At1g55300.1
[Arabidopsis thaliana] gi|25405812|pir||A96595
hypothetical protein F7A10.3 [imported] - Arabidopsis
thaliana gi|12323163|gb|AAG51559.1|AC027034_5
Serine/arginine-rich protein, putative; 48931-50251
[Arabidopsis thaliana]
Length = 203
Score = 171 bits (434), Expect = 7e-42
Identities = 92/149 (61%), Positives = 116/149 (77%), Gaps = 7/149 (4%)
Frame = -1
Query: 710 SLLDL-CVVESYKTYDDNSLIKSADIGQMIMVRESGDAAPDEIEYRHGLTPPMRDARKRR 534
SLLDL VVES+KTYDD +L+K+ADIGQMIMVRE GD AP+ +EYRHGLTPPM+DARKRR
Sbjct: 56 SLLDLPAVVESFKTYDDCALVKTADIGQMIMVREPGDPAPNTVEYRHGLTPPMKDARKRR 115
Query: 533 FRREPDLNPELVSRVEKDLLKIMAGGTADSFDVVAAEQEGDENVRGANKK---TAPTPEP 363
FRREPDLNPELV RVE+DLL I++GGT ++ E +EN A+KK ++PTP
Sbjct: 116 FRREPDLNPELVQRVERDLLNILSGGTVENVH-EQEEPATNENASNASKKVSSSSPTPVE 174
Query: 362 KHDAPETLTN---AGDPDRSDSDESDDSV 285
K +APET T+ +P+RS+S++SDDS+
Sbjct: 175 KPEAPETGTSNPTGVEPERSESEDSDDSM 203
>dbj|BAC00616.1| OJ1316_H05.12 [Oryza sativa (japonica cultivar-group)]
Length = 440
Score = 107 bits (268), Expect = 1e-22
Identities = 54/68 (79%), Positives = 60/68 (87%), Gaps = 1/68 (1%)
Frame = -1
Query: 710 SLLDL-CVVESYKTYDDNSLIKSADIGQMIMVRESGDAAPDEIEYRHGLTPPMRDARKRR 534
SLLDL V+ESYKTY+D+ LIK+ADIGQMIMVRE D AP+ IEY HGLTPPMRDAR+RR
Sbjct: 334 SLLDLPTVMESYKTYNDSVLIKTADIGQMIMVREEDDPAPEGIEYTHGLTPPMRDARRRR 393
Query: 533 FRREPDLN 510
FRREPDLN
Sbjct: 394 FRREPDLN 401
>ref|NP_499647.1| TBP-Associated transcription Factor family TAF-7.2 (26.4 kD)
(taf-7.2) [Caenorhabditis elegans]
gi|13548468|emb|CAC35845.1| C. elegans TAF-7.2 protein
(corresponding sequence Y111B2A.16) [Caenorhabditis
elegans]
Length = 236
Score = 60.5 bits (145), Expect = 2e-08
Identities = 48/160 (30%), Positives = 78/160 (48%), Gaps = 21/160 (13%)
Frame = -1
Query: 707 LLDL-CVVESYKTYDDNSLIKSADIGQMIM----------------VRESGDAAPDEIEY 579
+LDL + E +KT D+ SL K AD+ Q+++ +++ A + +Y
Sbjct: 88 ILDLPTITEIHKTLDNKSLYKVADVSQILVCTHDSINSIASSSEDAAQKAAAAKAKQWQY 147
Query: 578 RHGLTPPMRDARKRRFRREPDLNPELVSRVEKDLLKIM-AGGTADS--FDVVAAEQEG-D 411
HGLTPPM+ ARK+RFR+ VEK+L +++ A ADS +++V +EG
Sbjct: 148 PHGLTPPMKSARKKRFRKTKKKKFMDAPEVEKELKRLLRADLEADSVKWEIVEGNKEGAT 207
Query: 410 ENVRGANKKTAPTPEPKHDAPETLTNAGDPDRSDSDESDD 291
+ VR T P+ ++ + D D D+ DD
Sbjct: 208 DEVRTQRHVTYPS-----------SSEDESDVGDDDDKDD 236
>gb|EAA09978.1| agCP11819 [Anopheles gambiae str. PEST]
Length = 431
Score = 57.8 bits (138), Expect = 1e-07
Identities = 45/178 (25%), Positives = 80/178 (44%), Gaps = 34/178 (19%)
Frame = -1
Query: 692 VVESYKTYDDNSLIKSADIGQMIMVRESGDAAPDEIE------------------YRHGL 567
++ES KT D + K+ADI QM++ +E + E E + HG+
Sbjct: 90 IIESLKTIDSKNFYKTADICQMMICKEEPEQQSAEEESPNKNKKKDPNKVDKKFLWPHGV 149
Query: 566 TPPMRDARKRRFRREPDLN----PELVSRVEKDLLKIMAGGTADSFDVVAAEQEGDENVR 399
TPP ++ RKRRFR+ PE+ V K LL++ ++++ +++ ++
Sbjct: 150 TPPCKNIRKRRFRKTLKKKYVEAPEIEKEV-KRLLRVDNEAVNVKWELITEDEDPNKPSP 208
Query: 398 GANKKTAPTPEPKHDAPETLTNAGDPDR------------SDSDESDDSV*PKLAINE 261
G+ + P H +P T GD ++ SDSDE D+ + + ++E
Sbjct: 209 GSGNEAG----PPHKSPSKNTKKGDHNKDVGEHDIFGEEVSDSDEEDNPINKNIDLDE 262
>gb|EAA34629.1| hypothetical protein [Neurospora crassa]
Length = 563
Score = 57.0 bits (136), Expect = 3e-07
Identities = 49/151 (32%), Positives = 72/151 (47%), Gaps = 13/151 (8%)
Frame = -1
Query: 707 LLDL-CVVESYKTYDDNSLIKSADIGQMIMV----RESGDAA----PDEIEYRH----GL 567
L+DL + E KT+D S++KSADI QMI+V ++ +A P +E+ H G+
Sbjct: 261 LVDLPTITEGMKTWDKKSMVKSADICQMILVFAKVKDEAEAKSVPLPKAVEHGHRWPHGI 320
Query: 566 TPPMRDARKRRFRREPDLNPELVSRVEKDLLKIMAGGTADSFDVVAAEQEGDENVRGANK 387
TPPM DAR RRFR+ L+ + E ++ +++A D A + G +
Sbjct: 321 TPPMHDARNRRFRKR--LSKLEIQNKEAEVERLLAA------DREALNSRYELVAEGGHG 372
Query: 386 KTAPTPEPKHDAPETLTNAGDPDRSDSDESD 294
TA APE+ GD D E D
Sbjct: 373 NTA--------APESDHEYGDEDGGMGGEED 395
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 552,840,850
Number of Sequences: 1393205
Number of extensions: 11572675
Number of successful extensions: 36499
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 34205
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36282
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32654539052
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)