Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003416A_C01 KMC003416A_c01
(515 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
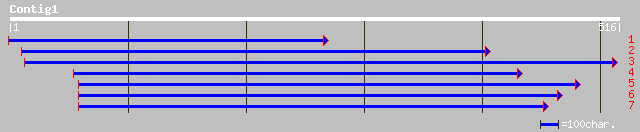
ref|NP_564333.1| expressed protein; protein id: At1g29690.1, sup... 154 8e-37
dbj|BAB56041.1| P0481E12.26 [Oryza sativa (japonica cultivar-gro... 148 4e-35
dbj|BAC07382.1| OJ1641_C04.15 [Oryza sativa (japonica cultivar-g... 80 1e-14
dbj|BAB88998.1| P0401G10.6 [Oryza sativa (japonica cultivar-group)] 79 4e-14
pir||T09892 hypothetical protein T22A6.120 - Arabidopsis thalian... 76 2e-13
>ref|NP_564333.1| expressed protein; protein id: At1g29690.1, supported by cDNA:
gi_15809819, supported by cDNA: gi_18650617 [Arabidopsis
thaliana] gi|25333974|pir||C86420 unknown protein,
124288-121737 [imported] - Arabidopsis thaliana
gi|12323548|gb|AAG51760.1|AC068667_39 unknown protein;
124288-121737 [Arabidopsis thaliana]
gi|18650618|gb|AAL75908.1| At1g29690/F15D2_24
[Arabidopsis thaliana]
Length = 561
Score = 154 bits (388), Expect = 8e-37
Identities = 75/113 (66%), Positives = 87/113 (76%)
Frame = -3
Query: 513 WDFGAQNVLHLKLLFSKVPGCTIRRSVWDHNPSTPATAAQRSDGASSSLTKKTSDDKKED 334
WDFG++NVLHLKLLFSKVPGCTIRRSVWDH P + + ++SS T++ S
Sbjct: 457 WDFGSKNVLHLKLLFSKVPGCTIRRSVWDHTPVASSGRLEPGGPSTSSSTEEVSGQS--- 513
Query: 333 SSIHIGKLAKIVDMTEMFRGPQDIPGHWLVTGAKLGVEKGKIVLRIKYSLLNY 175
GKLAKIVD +EM +GPQD+PGHWLVTGAKLGVEKGKIVLR+KYSLLNY
Sbjct: 514 -----GKLAKIVDSSEMLKGPQDLPGHWLVTGAKLGVEKGKIVLRVKYSLLNY 561
>dbj|BAB56041.1| P0481E12.26 [Oryza sativa (japonica cultivar-group)]
Length = 553
Score = 148 bits (373), Expect = 4e-35
Identities = 77/113 (68%), Positives = 88/113 (77%)
Frame = -3
Query: 513 WDFGAQNVLHLKLLFSKVPGCTIRRSVWDHNPSTPATAAQRSDGASSSLTKKTSDDKKED 334
WDFGA++VLHLKLLFS+VPGCTIRRSVWDH+PS+ + R+D ASSS +SD+
Sbjct: 456 WDFGAKSVLHLKLLFSRVPGCTIRRSVWDHSPSS--SLVHRTDEASSS----SSDN---- 505
Query: 333 SSIHIGKLAKIVDMTEMFRGPQDIPGHWLVTGAKLGVEKGKIVLRIKYSLLNY 175
KL KIVDMTE +GPQD PGHWLVTGAKLGVEKGKIV+R KYSLLNY
Sbjct: 506 -----AKLVKIVDMTETLKGPQDAPGHWLVTGAKLGVEKGKIVVRAKYSLLNY 553
>dbj|BAC07382.1| OJ1641_C04.15 [Oryza sativa (japonica cultivar-group)]
gi|22202779|dbj|BAC07435.1| P0039H02.26 [Oryza sativa
(japonica cultivar-group)]
Length = 601
Score = 80.5 bits (197), Expect = 1e-14
Identities = 52/128 (40%), Positives = 74/128 (57%), Gaps = 20/128 (15%)
Frame = -3
Query: 504 GAQNVLHLKLLFSKVPGCT-IRRSVWDHNPSTPATAAQRSDGASSSLTKKTSD------- 349
G + +L L+L FSKV T +R W+ +P+ AQ+S S+ ++ S
Sbjct: 462 GFKKILFLRLHFSKVCNATSVRNPEWEGSPNL----AQKSGLISTLISTHFSTAAQKPAP 517
Query: 348 ---DKKEDSSIHIG---------KLAKIVDMTEMFRGPQDIPGHWLVTGAKLGVEKGKIV 205
D +S+++ G KL K VD TEM RGPQD+PG+W+V+GAKL +E+GKI
Sbjct: 518 RPADVNINSAVYPGGPPVPVQTPKLLKFVDPTEMMRGPQDLPGYWVVSGAKLQLERGKIS 577
Query: 204 LRIKYSLL 181
LR+KYSLL
Sbjct: 578 LRVKYSLL 585
>dbj|BAB88998.1| P0401G10.6 [Oryza sativa (japonica cultivar-group)]
Length = 573
Score = 78.6 bits (192), Expect = 4e-14
Identities = 46/124 (37%), Positives = 67/124 (53%), Gaps = 16/124 (12%)
Frame = -3
Query: 504 GAQNVLHLKLLFSKVPGCT-IRRSVWDHNPSTPATAAQRSDGASSSLTKKTSDDKKEDSS 328
G + +L L+L FSKV T ++ WD +P+ + S S+ + +
Sbjct: 435 GFKKILFLRLHFSKVCNATSVKNPEWDGSPNLGQKSGLISTLISTHFSTAALKPAPRPAE 494
Query: 327 IHIG---------------KLAKIVDMTEMFRGPQDIPGHWLVTGAKLGVEKGKIVLRIK 193
++I KL + VD TEM RGPQD+PG+W+V+GAKL +E+GKI LR+K
Sbjct: 495 VNINSAVYPGGPPVPVQTPKLLRFVDTTEMLRGPQDLPGYWVVSGAKLHLERGKISLRVK 554
Query: 192 YSLL 181
YSLL
Sbjct: 555 YSLL 558
>pir||T09892 hypothetical protein T22A6.120 - Arabidopsis thaliana
gi|5051771|emb|CAB45064.1| putative protein [Arabidopsis
thaliana] gi|7269279|emb|CAB79339.1| putative protein
[Arabidopsis thaliana]
Length = 606
Score = 76.3 bits (186), Expect = 2e-13
Identities = 50/122 (40%), Positives = 67/122 (53%), Gaps = 15/122 (12%)
Frame = -3
Query: 504 GAQNVLHLKLLFSKVPGCT-IRRSVWDHNPSTPATAAQRSDGASSSLTKKTSD-----DK 343
G +NVL L+L FS+V G T ++ S WD + S S T D
Sbjct: 469 GFKNVLFLRLCFSRVVGATLVKNSEWDEAVGFAPKSGLISTLISHHFTAAQKPPPRPADV 528
Query: 342 KEDSSIHIG---------KLAKIVDMTEMFRGPQDIPGHWLVTGAKLGVEKGKIVLRIKY 190
+S+I+ G KL K VD +EM RGPQ+ PG+W+V+GA+L VEKGKI L++KY
Sbjct: 529 NINSAIYPGGPPVPTQAPKLLKFVDTSEMTRGPQESPGYWVVSGARLLVEKGKISLKVKY 588
Query: 189 SL 184
SL
Sbjct: 589 SL 590
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 425,249,922
Number of Sequences: 1393205
Number of extensions: 8614942
Number of successful extensions: 20865
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 20181
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20837
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16154357632
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)