Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003412A_C03 KMC003412A_c03
(634 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
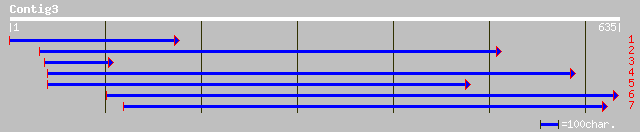
gb|ZP_00098281.1| hypothetical protein [Desulfitobacterium hafni... 36 0.48
gb|AAB41263.3| rhoptry protein [Plasmodium yoelii] 32 9.1
gb|EAA18154.1| rhoptry protein [Plasmodium yoelii yoelii] 32 9.1
pir||T28676 rhoptry protein - Plasmodium yoelii (fragment) 32 9.1
ref|NP_569959.1| CG3810-PB [Drosophila melanogaster] gi|24639212... 32 9.1
>gb|ZP_00098281.1| hypothetical protein [Desulfitobacterium hafniense]
Length = 473
Score = 35.8 bits (81), Expect = 0.48
Identities = 16/40 (40%), Positives = 23/40 (57%)
Frame = +2
Query: 380 SQMDNQRSTLVEL*IFGAQLSPLKSLVHSTHNLFHQFEQG 499
+++ N++ST+ IFG L PL H HNLFH +G
Sbjct: 343 AKLSNKKSTIQNFAIFGYSLIPLDLAGHIAHNLFHLLAEG 382
>gb|AAB41263.3| rhoptry protein [Plasmodium yoelii]
Length = 2771
Score = 31.6 bits (70), Expect = 9.1
Identities = 27/125 (21%), Positives = 53/125 (41%), Gaps = 21/125 (16%)
Frame = +2
Query: 17 INYIKSHVQNNINIF*KIITQMCQQQNN------TKPMSLNKFV*KY*LLL-----RTHS 163
IN IKS NN N+ + ++ + NN T LN + K ++ +
Sbjct: 2577 INLIKSEYNNNDNVSYNVAKKLEEDANNIIRDLDTSHNMLNDLIQKNFKIIDDLKNKKQE 2636
Query: 164 LKNLNQISYIS*KHELIMTKHAAGT----------TTNSNEVDSSSATT*NHQSLAVHTR 313
++N N + I+ + E+ T+H T + N+++ ++ NHQ+ + +
Sbjct: 2637 IENRNNLQTINREQEITQTEHVNNTYHHDINDINDVIDINDINDTNDINQNHQNSSSDKK 2696
Query: 314 QNSLT 328
+S T
Sbjct: 2697 DSSKT 2701
>gb|EAA18154.1| rhoptry protein [Plasmodium yoelii yoelii]
Length = 2070
Score = 31.6 bits (70), Expect = 9.1
Identities = 27/125 (21%), Positives = 53/125 (41%), Gaps = 21/125 (16%)
Frame = +2
Query: 17 INYIKSHVQNNINIF*KIITQMCQQQNN------TKPMSLNKFV*KY*LLL-----RTHS 163
IN IKS NN N+ + ++ + NN T LN + K ++ +
Sbjct: 1876 INLIKSEYNNNDNVSYNVAKKLEEDANNIIRDLDTSHNMLNDLIQKNFKIIDDLKNKKQE 1935
Query: 164 LKNLNQISYIS*KHELIMTKHAAGT----------TTNSNEVDSSSATT*NHQSLAVHTR 313
++N N + I+ + E+ T+H T + N+++ ++ NHQ+ + +
Sbjct: 1936 IENRNNLQTINREQEITQTEHVNNTYHHDINDINDVIDINDINDTNDINQNHQNSSSDKK 1995
Query: 314 QNSLT 328
+S T
Sbjct: 1996 DSSKT 2000
>pir||T28676 rhoptry protein - Plasmodium yoelii (fragment)
Length = 2401
Score = 31.6 bits (70), Expect = 9.1
Identities = 27/125 (21%), Positives = 53/125 (41%), Gaps = 21/125 (16%)
Frame = +2
Query: 17 INYIKSHVQNNINIF*KIITQMCQQQNN------TKPMSLNKFV*KY*LLL-----RTHS 163
IN IKS NN N+ + ++ + NN T LN + K ++ +
Sbjct: 2207 INLIKSEYNNNDNVSYNVAKKLEEDANNIIRDLDTSHNMLNDLIQKNFKIIDDLKNKKQE 2266
Query: 164 LKNLNQISYIS*KHELIMTKHAAGT----------TTNSNEVDSSSATT*NHQSLAVHTR 313
++N N + I+ + E+ T+H T + N+++ ++ NHQ+ + +
Sbjct: 2267 IENRNNLQTINREQEITQTEHVNNTYHHDINDINDVIDINDINDTNDINQNHQNSSSDKK 2326
Query: 314 QNSLT 328
+S T
Sbjct: 2327 DSSKT 2331
>ref|NP_569959.1| CG3810-PB [Drosophila melanogaster] gi|24639212|ref|NP_726777.1|
CG3810-PA [Drosophila melanogaster]
gi|24639214|ref|NP_726778.1| CG3810-PC [Drosophila
melanogaster] gi|7290228|gb|AAF45690.1| CG3810-PB
[Drosophila melanogaster] gi|7290229|gb|AAF45691.1|
CG3810-PC [Drosophila melanogaster]
gi|7290230|gb|AAF45692.1| CG3810-PA [Drosophila
melanogaster] gi|21429070|gb|AAM50254.1| LD21416p
[Drosophila melanogaster] gi|21711649|gb|AAM75015.1|
GH14693p [Drosophila melanogaster]
Length = 982
Score = 31.6 bits (70), Expect = 9.1
Identities = 17/60 (28%), Positives = 33/60 (54%)
Frame = -3
Query: 578 SAMMIYRANCNDKRITWSKPQWNTIKCPAQTDEIDCGYYVLSFLKEIVAHQKSTIPQAYF 399
SAM +YRA N+ + + + T++ A+T CGY + ++ +V H+K +++F
Sbjct: 382 SAMYLYRATGNEYLLEFGEHMLETLEFSAKT---KCGY---ATIRNVVTHEKENRMESFF 435
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 488,155,364
Number of Sequences: 1393205
Number of extensions: 9782610
Number of successful extensions: 20423
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 19984
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20421
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26154777244
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)